


 النبات
النبات
 الحيوان
الحيوان
 الأحياء المجهرية
الأحياء المجهرية
 علم الأمراض
علم الأمراض
 التقانة الإحيائية
التقانة الإحيائية
 التقنية الحيوية المكروبية
التقنية الحيوية المكروبية
 التقنية الحياتية النانوية
التقنية الحياتية النانوية
 علم الأجنة
علم الأجنة
 الأحياء الجزيئي
الأحياء الجزيئي
 علم وظائف الأعضاء
علم وظائف الأعضاء
 الغدد
الغدد
 المضادات الحيوية
المضادات الحيوية| Other Recoding Events: Translational Bypassing and the tmRNA Mechanism to Free Stalled Ribosomes |
|
|
|
Read More
Date: 7-5-2021
Date: 22-12-2015
Date: 16-5-2021
|
Other Recoding Events: Translational Bypassing and the tmRNA Mechanism to Free Stalled Ribosomes
KEY CONCEPTS
- Bypassing involves the capacity of the ribosome to stop translation, release from mRNA, and resume translation some 50 nucleotides downstream.
- Ribosomes that are stalled on mRNA after partial synthesis of a protein may be freed by the action of tmRNA, a unique RNA that incorporates features of both tRNA and mRNA.
Bypassing involves a movement of the ribosome to change the codon that is paired with the peptidyl-tRNA in the P site. The sequence between the two codons is skipped over and is not represented in the polypeptide product. As shown in FIGURE 1, this allows translation to continue past any termination codons in the intervening region. This is a very rare phenomenon; one of the few authenticated examples is that of gene 60 of phage T4, where the ribosome moves 60 nucleotides along the mRNA. Bypassing in individual cells has also been documented to be a result of nutrient starvation.
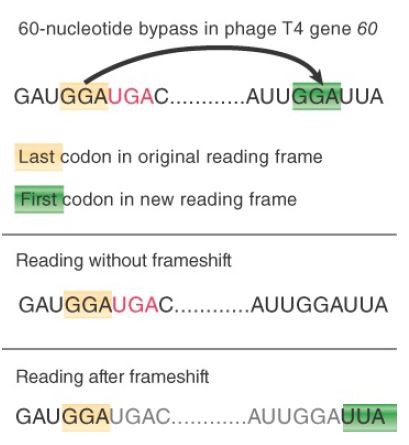
FIGURE 1.Bypassing occurs when the ribosome moves along mRNA so that the peptidyl-tRNA in the P site is released from pairing with its codon and then repairs with another codon farther along.
The key to the bypass system is that there are identical (or synonymous) codons at either end of the skipped sequence. These are sometimes referred to as the “takeoff” and “landing” sites. Before bypass, the ribosome is positioned with a peptidyl-tRNA paired with the takeoff codon in the P site, with an empty A site waiting for an aminoacyl-tRNA to enter. FIGURE 2 shows that the ribosome slides along mRNA in this condition until the peptidyltRNA can become paired with the codon in the landing site.

FIGURE 1. In bypass mode, a ribosome with its P site occupied can stop translation. It slides along mRNA to a site where peptidyl-tRNA pairs with a new codon in the P site. Then translation is resumed.
The sequence of the mRNA triggers the bypass. The important features are the two GGA codons for takeoff and landing, the spacing between them, a stem-loop structure that includes the takeoff codon, and a stop codon positioned adjacent to the takeoff codon.
The takeoff stage requires the peptidyl-tRNA to unpair from its codon. This is followed by a movement of the mRNA that prevents it from re-pairing. Then the ribosome scans the mRNA until the peptidyl-tRNA can re-pair with the codon in the landing reaction. This is followed by the resumption of translation when aminoacyltRNA enters the A site in the usual way.
Like frameshifting, the bypass reaction depends on a pause by the ribosome. The probability that peptidyl-tRNA will dissociate from its codon in the P site is increased by delays in the entry of aminoacyltRNA into the A site. Starvation for an amino acid can trigger bypassing in bacterial genes because of the delay that occurs when there is no aminoacyl-tRNA available to enter the A site. In phage T4 gene 60, one role of mRNA structure may be to reduce the efficiency of termination, thus creating the delay that is needed for the takeoff reaction.
The rescue of stalled ribosomes in bacteria and some mitochondria is accomplished by means of a unique mRNA–tRNA hybrid, termed tmRNA, which contains two functional domains. One domain mimics part of tRNA , whereas the second domain encodes a short polypeptide. tmRNA is first aminoacylated by alanyl-tRNA synthetase (AlaRS). It is then bound by EF-Tu and subsequently used in a ternary complex at the A site of stalled ribosomes. Peptidyl transfer occurs on the ribosome to join alanine to the Cterminal end of the stalled nascent protein; simultaneously, the mRNA present on the ribosome is replaced by the second domain of tmRNA. tmRNA then functions as a template for the synthesis of 10 additional amino acids, after which a stop codon is present to terminate translation and release the protein. The newly added Cterminal sequence then acts as a tag for subsequent recognition by proteases, which degrade the truncated protein. tmRNA thus functions as a quality-control mechanism to recycle stalled ribosomes and to remove truncated proteins that might otherwise accumulate.



|
|
|
|
دراسة يابانية لتقليل مخاطر أمراض المواليد منخفضي الوزن
|
|
|
|
|
|
|
اكتشاف أكبر مرجان في العالم قبالة سواحل جزر سليمان
|
|
|
|
|
|
|
اتحاد كليات الطب الملكية البريطانية يشيد بالمستوى العلمي لطلبة جامعة العميد وبيئتها التعليمية
|
|
|