

النبات

مواضيع عامة في علم النبات

الجذور - السيقان - الأوراق

النباتات الوعائية واللاوعائية

البذور (مغطاة البذور - عاريات البذور)

الطحالب

النباتات الطبية


الحيوان

مواضيع عامة في علم الحيوان

علم التشريح

التنوع الإحيائي

البايلوجيا الخلوية


الأحياء المجهرية

البكتيريا

الفطريات

الطفيليات

الفايروسات


علم الأمراض

الاورام

الامراض الوراثية

الامراض المناعية

الامراض المدارية

اضطرابات الدورة الدموية

مواضيع عامة في علم الامراض

الحشرات


التقانة الإحيائية

مواضيع عامة في التقانة الإحيائية


التقنية الحيوية المكروبية

التقنية الحيوية والميكروبات

الفعاليات الحيوية

وراثة الاحياء المجهرية

تصنيف الاحياء المجهرية

الاحياء المجهرية في الطبيعة

أيض الاجهاد

التقنية الحيوية والبيئة

التقنية الحيوية والطب

التقنية الحيوية والزراعة

التقنية الحيوية والصناعة

التقنية الحيوية والطاقة

البحار والطحالب الصغيرة

عزل البروتين

هندسة الجينات


التقنية الحياتية النانوية

مفاهيم التقنية الحيوية النانوية

التراكيب النانوية والمجاهر المستخدمة في رؤيتها

تصنيع وتخليق المواد النانوية

تطبيقات التقنية النانوية والحيوية النانوية

الرقائق والمتحسسات الحيوية

المصفوفات المجهرية وحاسوب الدنا

اللقاحات

البيئة والتلوث


علم الأجنة

اعضاء التكاثر وتشكل الاعراس

الاخصاب

التشطر

العصيبة وتشكل الجسيدات

تشكل اللواحق الجنينية

تكون المعيدة وظهور الطبقات الجنينية

مقدمة لعلم الاجنة


الأحياء الجزيئي

مواضيع عامة في الاحياء الجزيئي


علم وظائف الأعضاء


الغدد

مواضيع عامة في الغدد

الغدد الصم و هرموناتها

الجسم تحت السريري

الغدة النخامية

الغدة الكظرية

الغدة التناسلية

الغدة الدرقية والجار الدرقية

الغدة البنكرياسية

الغدة الصنوبرية

مواضيع عامة في علم وظائف الاعضاء

الخلية الحيوانية

الجهاز العصبي

أعضاء الحس

الجهاز العضلي

السوائل الجسمية

الجهاز الدوري والليمف

الجهاز التنفسي

الجهاز الهضمي

الجهاز البولي


المضادات الميكروبية

مواضيع عامة في المضادات الميكروبية

مضادات البكتيريا

مضادات الفطريات

مضادات الطفيليات

مضادات الفايروسات

علم الخلية

الوراثة

الأحياء العامة

المناعة

التحليلات المرضية

الكيمياء الحيوية

مواضيع متنوعة أخرى

الانزيمات
Alternative Splicing Is a Rule, Rather Than an Exception, in Multicellular Eukaryotes
المؤلف:
JOCELYN E. KREBS, ELLIOTT S. GOLDSTEIN and STEPHEN T. KILPATRICK
المصدر:
LEWIN’S GENES XII
الجزء والصفحة:
16-5-2021
2083
Alternative Splicing Is a Rule, Rather Than an Exception, in Multicellular Eukaryotes
KEY CONCEPTS
- Specific exons or exonic sequences may be excluded or included in the mRNA products by using alternative splicing sites.
- Alternative splicing contributes to structural and functional diversity of gene products.
- Sex determination in Drosophila involves a series of alternative splicing events in genes encoding successive products of a pathway.
When an interrupted gene is transcribed into an RNA that gives rise to a single type of spliced mRNA, the assignment of exons and introns is unambiguous. However, the RNAs of most mammalian genes follow patterns of alternative splicing, which occurs when a single gene gives rise to more than one mRNA sequence. By largescale cDNA cloning and sequencing, it has become apparent that more than 90% of the genes expressed in mammals are alternatively spliced. Thus, alternative splicing is not just the result of mistakes made by the splicing machinery; it is part of the gene expression program that results in multiple gene products from a single gene locus.
Various modes of alternative splicing have been identified, including intron retention, alternative 5′ splice-site selection, alternative 3′ splice-site selection, exon inclusion or skipping, and mutually exclusive selection of the alternative exons, as summarized in FIGURE 1. A single primary transcript may undergo more than one mode of alternative splicing. The mutually exclusive exons are normally regulated in a tissue-specific manner. Adding to this complexity, in some cases the ultimate pattern of expression is also dictated by the use of different transcription start points or the generation of alternative 3′ ends.
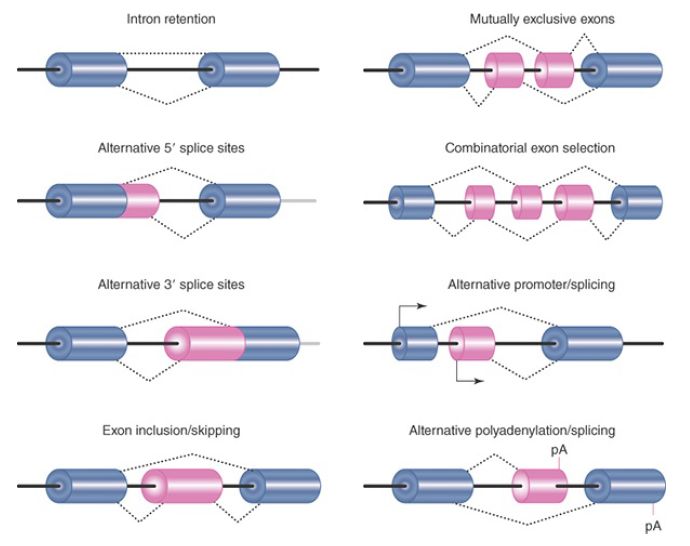
FIGURE 1. Different modes of alternative splicing.
Alternative splicing can affect gene expression in the cell in at least two ways. One way is to create structural diversity of gene products by including or omitting some coding sequences or by creating alternative reading frames for a portion of the gene. This can often modify the functional property of encoded proteins. For example, the CaMKIIδ gene contains three alternatively spliced exons, as shown in FIGURE 2. The gene is expressed in almost all cell types and tissues in mammals. When all three alternative exons are skipped, the mRNA encodes a cytoplasmic kinase that phosphorylates a large number of protein substrates.
When exon 14 is included, the kinase is transported to the nucleus because exon 14 contains a nuclear localization signal. This allows the kinase to regulate transcription in the nucleus. When both exons 15 and 16 are included, which is normally detected in neurons, the kinase is targeted to the cell membrane, where it can influence specific ion channel activities.
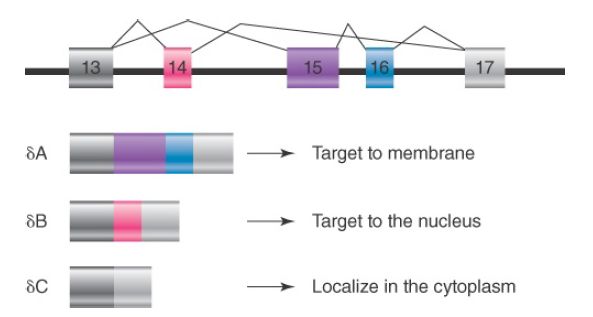
FIGURE 2. Alternative splicing of the CaMKIIδ gene: different alternative exons target the kinase to different cellular compartments.
In other cases, the alternatively spliced products exhibit opposite functions. This applies to essentially all genes involved in the regulation of apoptosis; each gene expresses at least two isoforms, one functioning to promote apoptosis and the other protecting cells against apoptosis. It is thought that the isoform ratios of these apoptosis regulators may dictate whether the cell lives or dies.
Alternative splicing may also affect various properties of the mRNA by including or omitting certain regulatory RNA elements, which may significantly alter the half-life of the mRNA. In many cases, the main purpose of alternative splicing may be to cause a certain percentage of primary transcripts to carry a premature stop codon(s) so that those transcripts can be rapidly degraded. This may represent an alternative strategy to transcriptional regulation to control the abundance of specific mRNAs in the cell. This mechanism is used to achieve homeostatic expression for many splicing regulators in specific cell types or tissues. In such regulation, a specific positive splicing regulator may affect its own alternative splicing, resulting in the inclusion of an exon containing a premature stop codon. This siphons a fraction of its mRNA to
degradation, thereby reducing the protein concentration. Thus, when the concentration of such positive splicing regulator fluctuates in the cell, its mRNA concentration will be shifted in the opposite direction.
Although many alternative splicing events have been characterized and the biological roles of the alternatively spliced products determined, the best understood example is still the pathway of sex determination in D. melanogaster, which involves interactions between a series of genes in which alternative splicing events distinguish males and females. The pathway takes the form
illustrated in FIGURE 3, in which the ratio of X chromosomes to autosomes determines the expression of sex lethal (sxl), and changes in expression are passed sequentially through the other genes to doublesex (dsx), the last in the pathway.
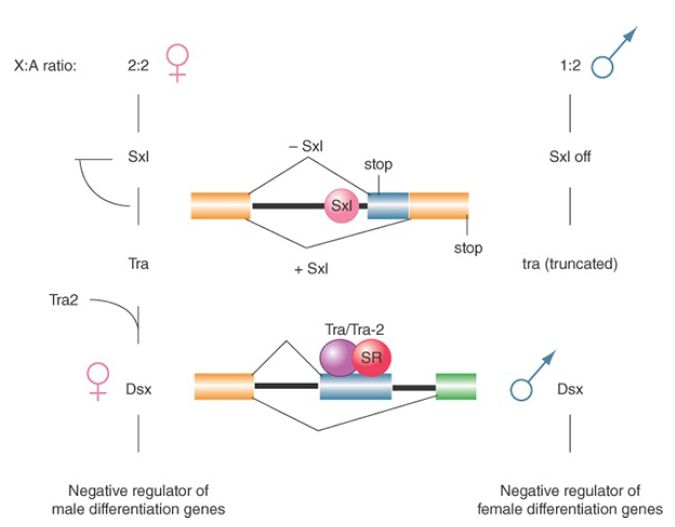
FIGURE 3. Sex determination in D. melanogaster involves a pathway in which different splicing events occur in females. Blockages at any stage of the pathway result in male development. Illustrated are tra pre-mRNA splicing controlled by the Sxl protein, which blocks the use of the alternative 3′ splice site, and dsx premRNA splicing regulated by both Tra and Tra2 proteins in conjunction with other SR proteins, which positively influence the inclusion of the alternative exon.
The pathway starts with sex-specific splicing of sxl. Exon 3 of the sxl gene contains a termination codon that prevents synthesis of functional protein. This exon is included in the mRNA produced in males but is skipped in females. As a result, only females produce Sxl protein. The protein has a concentration of basic amino acids that resembles other RNA-binding proteins. The presence of Sxl protein changes the splicing of the transformer (tra) gene. Figure 3 shows that this involves splicing a constant 5′ site to alternative 3′ sites (note that this mode applies to both sxl and tra splicing, as illustrated). One splicing pattern occurs in both males and females and results in an RNA that has an early termination codon. The presence of Sxl protein inhibits usage of the upstream 3′ splice site by binding to the polypyrimidine tract at its branch site. When this site is skipped, the next 3′ site is used. This generates a female-specific mRNA that encodes a protein.
Thus, Sxl autoregulates the splicing of its own mRNA to ensure its expression in females, and tra produces a protein only in females; like Sxl, Tra protein is a splicing regulator. tra2 has a similar function in females (but is also expressed in the males). The Tra and Tra2 proteins are SR splicing factors that act directly upon the target transcripts. Tra and Tra2 cooperate (in females) to affect the splicing of dsx. In the dsx gene, females splice the 5′ site of intron 3 to the 3′ site of that intron; as a result, translation terminates at the end of exon 4. Males splice the 5′ site of intron 3 directly to the 3′ site of intron 4, thus omitting exon 4 from the mRNA and allowing translation to continue through exon 6. The result of the alternative splicing is that different Dsx proteins are produced in each sex: The male product blocks female sexual differentiation, whereas the female product represses expression of male-specific genes.
 الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
 اخر الاخبار
اخر الاخبار
اخبار العتبة العباسية المقدسة

الآخبار الصحية















 قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة
قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة "المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة
"المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة (نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)
(نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)


















