


 النبات
النبات
 الحيوان
الحيوان
 الأحياء المجهرية
الأحياء المجهرية
 علم الأمراض
علم الأمراض
 التقانة الإحيائية
التقانة الإحيائية
 التقنية الحيوية المكروبية
التقنية الحيوية المكروبية
 التقنية الحياتية النانوية
التقنية الحياتية النانوية
 علم الأجنة
علم الأجنة
 الأحياء الجزيئي
الأحياء الجزيئي
 علم وظائف الأعضاء
علم وظائف الأعضاء
 الغدد
الغدد
 المضادات الحيوية
المضادات الحيوية|
Read More
Date: 28-4-2016
Date: 3-6-2021
Date: 15-11-2020
|
Abscisic Acid
1. History
The first evidence for the existence of an acidic inhibitor of coleoptile growth that promoted abscission and seed maturation dates back to the early 1950s, but it was not until 1963 that abscisic acid (ABA) was identified by Frederick Addicott and coworkers (1, 2). Addicott's team was studying compounds that stimulated abscission in cotton fruits and had named the active substances abscisin I and abscisin II; the latter proved to be ABA. Two other independent efforts also culminated in the discovery of ABA. A British group headed by Wareing (3) was investigating bud dormancy of woody plants and called the most active molecule dormin. In New Zealand, van Stevenick (4) studied compounds that accelerated abscission of flowers and fruits of Lupinus luteus. In 1964, it became evident that the three groups had discovered the same plant hormone, which was renamed abscisic acid 3 years later.
2. Biosynthesis and Metabolism
ABA is a universal compound in vascular plants; it is not found in bacteria, but has been reported in green algae, certain fungi, and mosses (5). ABA is a sesquiterpenoid, with mevalonic acid as a precursor. In certain fungi, ABA is produced by a direct, C15 pathway from farnesyl pyrophosphate. In higher plants, ABA is derived from xanthophylls, via an indirect C40 pathway (Fig. 1). Substantial progress in understanding the ABA biosynthetic pathway has been achieved by a combination of molecular and genetic techniques, primarily on mutants in Arabidopsis thaliana, Zea mays, Nicotiana plumbaginifolia, and tomato. Arabidopsis mutants impaired in ABA biosynthesis were isolated on the basis of their lack of seed dormancy due to ABA deficiency and their ability to overcome a gibberellin requirement for germination, which allowed them to germinate in the presence of inhibitors of gibberellin biosynthesis or in a gibberellin-deficient background (6, 7). ABA-deficient mutants show a wilty phenotype when subjected to water stress.
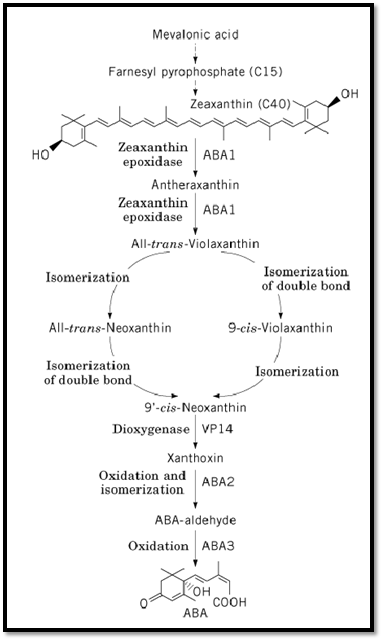
Figure 1. ABA biosynthesis in higher plants. Steps mediated by ABA1, ABA2, and ABA3 in Arabidopsis and by VP14 in Zea mays are indicated.
Mevalonic acid is converted to farnesyl pyrophosphate, a C15 compound, via several intermediates )Fig. 1). The subsequent conversion of farnesyl pyrophosphate to zeaxanthin, a C40 carotenoid, again involves multiple steps (8). A number of viviparous (vp) mutants of maize identify loci corresponding to these early conversions. The transformation of zeaxanthin to all-trans-violaxanthin consists of two epoxidations at the double bonds in both cyclohexenyl rings, with antheraxanthin as an intermediate. The aba1 mutant impaired in zeaxanthin epoxidase activity has been characterized biochemically in Arabidopsis (6, 9, 10). The N. plumbaginifolia ABA2 gene encoding zeaxanthin epoxidase was recently cloned by transposon tagging (11). The gene encodes a chloroplast-imported polypeptide chain with similarity to bacterial monooxygenases and oxidases. When the Nicotiana ABA2 gene was expressed in Escherichia coli heterologously, the protein was shown to catalyze both epoxidations in vitro. Moreover, the complementary DNA (cDNA) complemented both the N. plumbaginifolia aba2 mutant and the Arabidopsis aba1 mutant. The ortholog in Arabidopsis (ABA1) was cloned by homology with Nicotiana ABA2 (11). Although presumably a null allele, the Nicotiana aba2 mutant retains up to half the ABA content of wild type in the absence of water stress. This characteristic might indicate the existence of a secondary biosynthetic pathway for ABA. Downstream of all-trans-violaxanthin, two isomerizations yield 9′-cis-neoxanthin.
The subsequent oxidative cleavage with xanthoxin as a product is thought to be the rate-limiting step in ABA biosynthesis (12, 13). A gene that most probably encodes the enzyme performing this reaction was cloned from maize, and by homology also from Arabidopsis (VP14) (13). Its overproduction in E. coli indicated that VP14 is probably a member of a novel class of dioxygenases.
Xanthoxin is subsequently converted to ABA-aldehyde by oxidation and isomerization. Biochemical analysis indicates that the Arabidopsis aba2 mutant is blocked at this step in the pathway (7, 14). A final oxidation, catalyzed by ABA-aldehyde oxidase, produces ABA. Mutants in this step have been identified in several species (8). The Arabidopsis aba3 mutant lacks several aldehyde oxidase activities that require a molybdenum cofactor (14). Yet the mutation blocks modification of the mobybdenum cofactor, rather than impairing the activity of the apoprotein. Cloning of the ABA2 and ABA3 genes should provide further clues as to the regulation of ABA biosynthesis in these final steps.
Metabolism of ABA is mainly to phaseic acid, dihydrophaseic acid, and their respective conjugates (15, 16). Direct conjugation of ABA to an ABA glucose ester, or an ABA-b-glucopyranoside, can also occur. Conjugation of ABA does not lead to its storage, probably because it is irreversible and yields unstable compounds. With the exception of phaseic acid, none of the metabolites carries biological activity (16).
3. Signal Perception and Transduction
Insight into the molecular basis of ABA responses was gathered by a combination of molecular-genetic, biochemical, and electrophysiological studies (12, 17-20). Currently, little is known about the perception of ABA, despite numerous efforts to isolate ABA receptors using different approaches (20). Two groups of mutants, ABA-insensitive and ABA-hypersensitive, have largely contributed to the understanding of ABA signal transduction. Whereas the five abscisic acid-insensitive (abi) mutants of Arabidopsis were identified by their ability to germinate on inhibiting concentrations of ABA (21, 22), the three enhanced response to ABA (era) mutants showed an increased dormancy in the presence of low ABA concentrations, compared to wild type (23). Besides their effects on seed development and dormancy, these mutants show altered responses to drought adaptation. The abi mutants differ from the above-mentioned aba mutants, because they do not have reduced endogenous ABA content and their phenotype cannot be reverted by exogenously supplied ABA. The characterization of abi mutants revealed that at least two ABA-response pathways exist: one primarily active in vegetative tissues, involving ABI1 and ABI2, and a second one operating predominantly during seed development and involving ABI3, ABI4, and ABI5 (19). Genes corresponding to four loci have been cloned. ABI1 and ABI2 encode highly homologous serine/threonine kinases and phosphatases 2C (24-26). These phosphatases might act in a phosphorylation/dephosphorylation cascade, as the protein phosphatase activity of recombinant ABI1 has been demonstrated in vitro (27). The functions of ABI1 and ABI2 are partially redundant (25). ABI3 encodes a putative transcription factor that acts mainly during seed development and is the ortholog of VP1 in maize (28-30). The suggested function of ABI3 consists of either activating the maturation program or preventing germination (30, 31). Finally, the predicted amino acid sequence of ERA1 shares similarity with the b-subunit of farnesyl transferases. In yeast and mammalian systems, this enzyme is known to modify signal transduction proteins for membrane anchoring (23). In wild-type Arabidopsis, ERA1 is proposed to modify a negative regulator of ABA signaling.
The current model of ABA signal transduction integrates ABI1/ABI2-dependent cascades in both seeds and vegetative tissues (Fig. 2). These cascades interact with the ABI3 protein in seeds only, because ABI3 is expressed exclusively in seeds (32). The role of ABI3 is not confined to ABA signaling (19); it is also thought to mediate developmental signals. This role is supported by the seed phenotype of abi3, which is more complex than that of abi1, abi2, or aba1 mutants (19). The evidence that VP1 can also regulate transcription independently of ABA further supports the possibility of a less strict limitation of ABI3 to ABA signaling (33, 34). The same holds true for ABI4 and ABI5 (2).
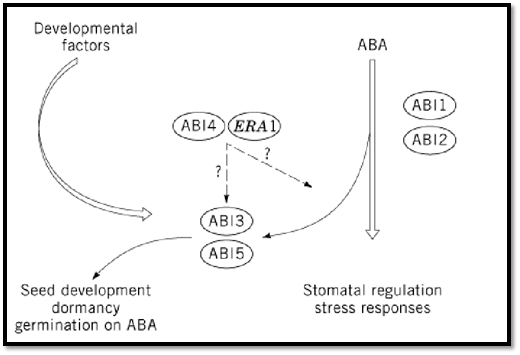
Figure 2. A model for ABA signal transduction.
Intermediates in the ABA transduction chain were revealed by microinjection of putative second messengers into hypocotyl cells of the aurea mutant of tomato (35). Following injection of ABA-inducible KIN2 and RD29A promoters linked to b-glucuronidase, coinjection of either Ca2+, cyclic ADP-ribose (cADP), ADP-ribosyl cyclase, or inositol (1,4,5)-triphosphate (IP3) resulted in expression of the ABA-inducible genes in the absence of ABA, implying that cADP ribose, a Ca2+-mobilizing second messenger, is involved in ABA responses. At present, it is difficult to integrate these data into the current model for the ABA signal transduction pathway (19). Electrophysiological studies (voltage clamp or patch clamp) on the ABA-insensitive mutants abi1 and abi2 have clearly shown an altered ion channel behavior in their stomatal guard cells and provided further evidence that phosphorylation/dephosphorylation cascades are involved in ABA signaling (36, 37(.
Given the power of mutational approaches to identify signal transduction components, several groups are currently aiming at the identification of additional ABA signal transduction factors, exploiting novel screening procedures in Arabidopsis (12). At least eight growth control via ABA (gca) loci have been identified on the basis of their insensitivity to ABA inhibition of root growth (38). Furthermore, a freezing sensitive (frs1) mutant with a wilty phenotype appeared to be nonallelic to other ABA-deficient or -insensitive mutants of Arabidopsis (39). Other screens employ transgenic lines carrying chimeric promoter-luciferase constructs with promoters derived from ABA-responsive genes. Putative mutants are identified by aberrant expression of these genes in the mutagenized progeny (40). The identification of intragenic or extragenic enhancers and suppressors of known signaling components is yet another alternative.
Whereas part of the ABA signaling pathway is well established in seeds and stomatal guard cells, the implications of ABA signaling in the function of the vegetative meristem remain to be clarified. Furthermore, it will be most interesting to characterize the regulatory influence of light on ABA signaling, as well as the crosstalk with gibberellins, which are known to counteract ABA in many physiological processes (5).
4. Downstream Targets
Of all the plant hormones, ABA is probably best known from the point of view of responsiveness at the target gene level. ABA-responsive genes encode proteins with diverse functions and are often induced under water stress (drought, salinity, low temperature). It is important to mention, however, that certain water stress-related genes are independent of ABA (17, 18). ABA-responsive genes can be classified into two groups. Primary ABA response genes are rapidly induced and are independent of protein biosynthesis, implying that trans-acting factors controlling these genes are under post-translational control. A second class of ABA response genes consists of those that require the expression of other genes.
Cis-acting and trans-acting elements involved in ABA induction of gene expression have been studied extensively, often in relation to water stress (17, 18). A bipartite model for regulation of transcription of the primary response genes is proposed, as for the barley HVA1 and HVA22 genes (41, 42). The specificity of ABA responsiveness relies on the combination of an ABA response element (ABRE) and a coupling element. The ABRE has a consensus sequence (PyACGTGGC) that contains a G-box core sequence and has been shown to interact with bZIP factors (eg, wheat Em gene-binding protein EmBP-1) (43). A set of unique coupling elements would provide specificity in ABA response at the level of individual genes. In addition, in the case of Em induction, it has been demonstrated that a conserved domain of 18 amino acid residues of the VP1 protein enhances DNA-binding activity of the EmBP-1 bZIP factor, thereby acting as a DNA chaperone (44). A yeast two-hybrid system screen was used to identify proteins that interact specifically with VP1 on the promoter of the Em gene. One such protein was related to 14–3–3 proteins (45) and thus may help to stabilize and/or activate the regulatory complex at the Em promoter (20(.
ABREs are not the only cis elements that confer ABA responsiveness. Both in the case of seed desiccation and in water stress conditions, other promoter elements can be involved. For example, ABA responses can be mediated in conjunction with the Sph box in germination-specific promoters. A direct interaction between the Sph box and a 140-residue conserved domain in the C-terminal region of VP1 has been demonstrated (46).
The second pathway for ABA-dependent gene expression requires protein biosynthesis, exemplified by the Arabidopsis ABA-inducible RD22 gene. A 67-bp region in the promoter of this gene contains conserved motifs for MYC and MYB factor binding (47) (see Oncogenes, Oncoproteins). Factors binding to these promoter elements (RD22BP-1 and ATMYB2) were shown to activate transcription of RD22 in transient transactivation assays (48). Cooperative binding of these factors has been proposed to control ABA-dependent expression of RD22.
One of the challenges for the future consists in making the link between the ABA-induced proteins and the tissue- and development-specific physiological responses, summarized below.
5. Effects
Although abscission was originally considered to be a process regulated by ABA, and the hormone was named accordingly, it is now clear that ethylene is the primary factor controlling abscission. Nevertheless, ABA also plays a key role in such diverse plant growth and developmental processes as seed maturation, germination, root growth, and stomatal closure (5). Stomatal movements involve changes in ion fluxes that occur very rapidly in response to ABA and do not require gene expression. Furthermore, ABA is a major stress signal orchestrating responses to dehydration stress, including water stress induced by drought and high-salt concentration as well as by low-temperature conditions.
Refernces
1. T. A. Bennet-Clark and N. P. Kefford (1953) Nature 171, 645–647.
2. K. Ohkuma, O. E. Smith, J. L. Lyon, and F. T. Addicott (1963) Science 142, 1592–1593.
3. P. F. Wareing (1956) Annu. Rev. Plant Physiol. 7, 191–214.
4. R. F. M. van Stevenick (1957) J. Exp. Bot. 8, 373–381.
5. P. J. Davies (1995) Plant Hormones: Physiology, Biochemistry and Molecular Biology, Kluwer, Dordrecht, The Netherlands.
6. M. Koornneef, M. L. Jorna, D. L. C. Brinkhorst-van der Swan, and C. M. Karssen (1982) Theor. Appl. Genet. 61, 385–393.
7. K. M. Léon-Kloosterziel, M. Alvarez Gil, G. J. Ruijs, S. E. Jacobsen, N. E. Olszewski, S. H. Schwartz, J. A. D. Zeevaart, and M. Koornneef (1996) Plant J. 10, 655–661.
8. I. B. Taylor (1991) In Abscisic Acid, Physiology and Biochemistry (W. J. Davies and H. G. Jones, eds.), Bios Scientific, Oxford, UK, pp. 23–37.
9. S. C. Duckham, R. S. T. Linforth, and I. B. Taylor (1991) Plant Cell Environ. 14, 631–636.
10. C. D. Rock and J. A. D. Zeevaart (1991) Proc. Natl. Acad. Sci. USA 88, 7496–7499.
11. E. Marin, L. Nussaume, A. Quesada, M. Gonneau, B. Sotta, P. Hugueney, A. Frey, and A. Marion-Poll (1996) EMBO J. 15, 2331–2342.
12. M. Koornneef, K. M. Léon-Kloosterziel, S. H. Schwartz, and J. A. D. Zeevaart (1998) Plant Physiol. Biochem. 36, 83–89.
13. S. H. Schwartz, K. M. Léon-Kloosterziel, M. Koornneef, and J. A. D. Zeevaart (1997) Plant Physiol. 114, 161–166.
14. S. H. Schwartz, B. C. Cai, D. A. Gage, J. A. D. Zeevaart, and D. R. McCarty (1997) Science 276, 1872–1874.
15. J. A. D. Zeevaart and R. A. Creelman (1988) Annu. Rev. Plant Physiol. Plant Mol. Biol. 39, 439–473.
16. G. Sembdner, R. Atzorn, and G. Schneider (1994) Plant Mol. Biol. 26, 1459–1481.
17. J. Ingram and D. Bartels (1996) Annu. Rev. Plant Physiol. Plant Mol. Biol. 47, 377–403.
18. K. Shinozaki and K. Yamaguchi-Shinozaki (1996) Curr. Opin. Biotechnol. 7, 161–167.
19. S. Merlot and J. Giraudat (1997) Plant Physiol. 114, 751–757.
20. R. S. Quatrano, D. Bartels, T. H. D. Ho, and M. Pagés (1997) Plant Cell 9, 470–475.
21. M. Koornneef, G. Reuling, and C. M. Karssen (1984) Physiol. Plant. 61, 377–383.
22. R. R. Finkelstein (1994) Plant J. 5, 765–771.
23. S. Cutler, M. Ghassemian, D. Bonetta, S. Cooney, and P. McCourt (1996) Science 273, -1241 1239.
24. J. Leung, M. Bouvier-Durand, P.-C. Morris, D. Guerrier, F. Chefdor, and J. Giraudat (1994). Science 264, 1448–1452.
25. J. Leung, S. Merlot, and J. Giraudat (1997) Plant Cell 9, 759–771.
26. K. Meyer, M. P. Leube, and E. Grill (1994) Science 264, 1452–1455
27 . N. Bertauche, J. Leung, and J. Giraudat (1996) Eur. J. Biochem. 241, 193–200.
28. D. R. McCarty, T. Hattori, C. B. Carson, V. Vasil, M. Lazar, and I. K. Vasil (1991) Cell 66 895–905,
29 J. Giraudat, B. M. Hauge, C. Valon, J. Smalle, F. Parcy, and H. M. Goodman (1992) Plant Cell 1251–1261 4,
30. F. Parcy, C. Valon, M. Raynal, P. Gaubier-Comella, M. Delseny, and J. Giraudat (1994) Plant Cell 6, 1567–1582.
31. E. Nambara, K. Keith, P. McCourt, and S. Naito (1995) Development 121, 629–636.
32. F. Parcy and J. Giraudat (1997) Plant J. 11, 693–702.
33. V. Vasil, W. R. Marcotte Jr., L. Rosenkrans, S. M. Cocciolone, I. K. Vasil, R. S. Quatrano, and D. R. McCarty (1995) Plant Cell 7, 1511–1518.
34. C.-Y. Kao, S. M. Cocciolone, I. K. Vasil, and D. R. McCarty (1996) Plant Cell 8, 1171–1179.
35. Y. Wu, J. Kuzma, E. Maréchal, R. Graeff, H. C. Lee, R. Foster, and C.-H. Hua (1997) Science. 2126–2130278,
36. F. Armstrong, J. Leung, A. Grabov, J. Brearley, J. Giraudat, and M. R. Blatt (1995) Proc. Natl. Acad. Sci. USA 92, 9520–9524.
37. Z.-M. Pei, K. Kuchitsu, J. M. Ward, M. Schwarz, and J. I. Schroeder (1997) Plant Cell 9, 409423–
38. G. Benning, T. Ehrler, K. Meyer, M. Leube, P. Rodriguez, and E. Grill (1996) In Abscisic Acid Signal Transduction in Plants. Centro de Reuniones Internacionales sobre Biología Workshop 60 )R. S. Quatrano and M. Pagès, eds.), Instituto Juan March de Estudios e Investigaciones,
Madrid, p. 34.
39. J. Salinas, F. Llorente, and J. M. Martinez-Zapater (1996) In Abscisic Acid Signal Transduction in Plants. Centro de Reuniones Internacionales sobre Biología Workshop 60 (R. S. Quatrano and M. Pagès, eds.), Instituto Juan March de Estudios e Investigaciones, Madrid, p. 35.
40. N.-H. Chua, Y. Wu, and R. Foster (1996) In Abscisic Acid Signal Transduction in Plants. Centro de Reuniones Internacionales sobre Biología Workshop 60 (R. S. Quatrano and M. Pagès, eds.), Instituto Juan March de Estudios e Investigaciones, Madrid, p. 55.
41. Q. Shen and T.-H. D. Ho (1995) Plant Cell 7, 295–307.
42 . Q. Shen, P. Zhang, and T.-H. D. Ho (1996) Plant Cell 8, 1107–1119
43. M. J. Guiltinan, W. R. Marcotte Jr., and R. S. Quatrano (1990) Science 250, 267–271.
44. A. Hill, A. Nantel, C. D. Rock, and R. S. Quatrano (1996) J. Biol. Chem. 271, 3366–3374.
45. T. F. Schultz, J. Medina, A. Hill, and R. S. Quatrano (1998) Plant Cell 10, 837–847.
46. M. Suzuki, C. Y. Kao, and D. R. McCarty (1997) Plant Cell 9, 799–807.
47. T. Iwasaki, K. Yamaguchi-Shinozaki, and K. Shinozaki (1995) Mol. Gen. Genet. 247, 391–398.
48. H. Abe, K. Yamaguchi-Shinozaki, T. Urao, T. Iwasaki, D. Hosokawa, and K. Shinozaki (1997) Plant Cell 9, 1859–1868.



|
|
|
|
دراسة يابانية لتقليل مخاطر أمراض المواليد منخفضي الوزن
|
|
|
|
|
|
|
اكتشاف أكبر مرجان في العالم قبالة سواحل جزر سليمان
|
|
|
|
|
|
|
المجمع العلمي ينظّم ندوة حوارية حول مفهوم العولمة الرقمية في بابل
|
|
|