


 النبات
النبات
 الحيوان
الحيوان
 الأحياء المجهرية
الأحياء المجهرية
 علم الأمراض
علم الأمراض
 التقانة الإحيائية
التقانة الإحيائية
 التقنية الحيوية المكروبية
التقنية الحيوية المكروبية
 التقنية الحياتية النانوية
التقنية الحياتية النانوية
 علم الأجنة
علم الأجنة
 الأحياء الجزيئي
الأحياء الجزيئي
 علم وظائف الأعضاء
علم وظائف الأعضاء
 الغدد
الغدد
 المضادات الحيوية
المضادات الحيوية|
Read More
Date: 14-3-2021
Date: 21-4-2016
Date: 30-3-2021
|
Complementation Tests
The gene is best defined as an integral unit of genetic function, in the sense that none of its parts functions normally in isolation from the others. If so, two different forms (alleles) of the same gene, both more or less inactivated by mutation, should not make good each other's defects when together in the same cell. They should not, in other words, show complementation. On the other hand, two genomes that have mutations in different genes should complement each other to produce a wild-type phenotype, provided that each mutant allele is recessive to its wild-type counterpart. This complementation criterion is of great importance because it permits assigning mutations to the same or to different genes without a detailed analysis of gene functions. When mutations are closely linked and have fairly similar phenotypic effects, it is always likely that they are in the same gene. But it is not uncommon to find close linkages of separate genes whose function is related. A complementation test provides much stronger evidence.
The interpretation of complementation tests is itself subject to certain difficulties, which are discussed separately under the heading Allelic complementation. Here we make the simplifying assumption that complementation between recessive mutations means that they are in different genes.
For a complementation test, it is necessary to bring two mutant genomes, or at least relevant parts of genomes, together in the same cell. In organisms including animals, higher plants and budding yeasts, that have a stable diploid phase in their life cycles, this can be simply achieved by making a sexual cross between mutant strains. In microorganisms that have no stable diploid phase, various other means have to be employed. In haploid filamentous fungi, heterokaryons that have two genetically different kinds of nuclei in a common cytoplasm provide a satisfactory substitute for diploidy. In bacteria, where neither diploidy nor heterokaryosis is usually available, there are various ways of introducing fragments of the genome from one cell to another to make partial diploids (merodiploids). This makes it possible to test for complementation of genes located within the duplicated fragment (Fig. 1). In viruses, especially bacterial viruses (bacteriophages), complementation tests can be carried out simply by mixed infections.
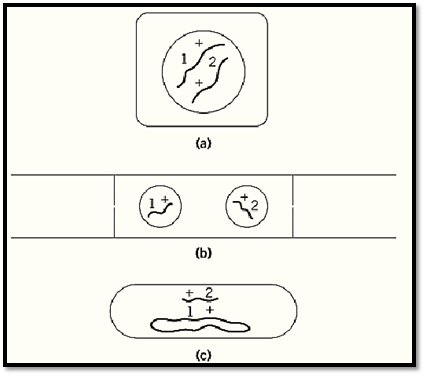
Figure 1. Three general ways of making complementation tests: (a) by diploidy (higher plants, animals and the budding yeast Saccharomyces cerevisiae); (b) by heterokaryosis (fungi, such as Neurospora crassa and sometimes cultured mammalian cells); (c) by addition of a genomic fragment to a haploid genome (bacteria, such as Escherichia coli and Salmonella typhimurium). In each diagram, 1 and 2 are different mutations, and + stands for the corresponding nonmutated sites.
1. Complementation Tests in Diploids
The most extensive complementation testing using diploids has been carried out in the budding yeast Saccharomyces cerevisiae and the fruit fly Drosophila melanogaster. These are the diploid organisms that have most experimentally induced mutants sorted into genes.
Saccharomyces has the great advantage as an experimental organism of propagating itself equally well as a haploid or a diploid. Haploid cells are of two different mating types, called a and a. In wild strains, the cells are constantly interconverted by a genetic switching mechanism but are stabilized in nonswitching laboratory strains. Most yeast genetics is based on the use of auxotrophic mutants that grow only if given some specific nutritional supplement which they can no longer synthesize—most commonly an amino acid, a purine or pyrimidine base, or a vitamin. Such mutants can be readily obtained in haploid strains. For comprehensive complementation testing, it is necessary to isolate each mutant in both mating types from a cross to wild type. Then haploid cells of pairs of mutants of opposite mating types are mixed at marked positions on plates of unsupplemented agar growth medium. Sexual fusion to form diploid cells occurs almost immediately, and the diploids grow if the two mutations complement each other's nutritional deficiencies. The result is quite clear after overnight incubation. Auxotrophs that have different nutritional requirements always complement, but mutants that have the same requirement may or may not do so because several different genes acting in sequence are usually necessary to synthesize a single end product. The results split the mutants that have a common requirement, say, for the amino acid histidine, into clear-cut complementation groups - his1, his2, his3, etc. Every member of one group complements every members of any other group, but only sporadic and fairly infrequent complementation occurs within groups. In general, complementation groups equate to genes, and each one is deficient in one enzyme of the biosynthetic pathway.
In the fruit-fly, Drosophila melanogaster, which is certainly the most intensively investigated multicellular organism, many mutants have been assigned to allelic series based on the mutant phenotypes of their heteroallelic combinations. The famous example of multiple alleles of the w (white-eye) gene is referred to under Complementation. Most of the allelic series have consisted of viable mutants, all within the same short chromosome segment, and obviously related phenotypes that made their allelic relationship are very likely even without complementation testing. However, nonvisible mutations, recessive lethals, have been much used in attempts to establish the total number of genes that have essential functions residing within a particular chromosome segment. Complementation is the only guide for classification of lethals.
A broadly applicable experimental scheme is shown in Fig. 2. The method is selecting, after heavy mutagenization, a large number of recessive lethal chromosomes based on their failure to complement a deletion of a short, defined chromosome segment, and maintaining them in breeding stocks combined with a “balancer” chromosome. A Drosophila balancer chromosome carries a dominant viable mutation to show that it is present and a recessive lethal mutation to ensure that any fly that carries it also has the chromosome against which the balancer is supposed to be balanced. The balancer chromosome also contains an inverted segment, or complex of inverted segments, to inhibit its recombination with the mutagenized chromosome, or else has genetic markers to ensure that recombination within the mutagenized region can be avoided. Crosses between flies that have different recessive lethals, each in combination with the same balancer, produce a predicted one-third of their progeny free of the balancer, if the two are in different genes and therefore complement one another, but no progeny without the balancer if they are in the same gene.

Figure 2. The isolation of recessive lethal mutations in Drosophila and testing them for complementation. (a) Male flies treated with a mutagen and mated to females carrying a balancer third chromosome that has the marker mutations Sb (dobristle phenotype, recessive lethal) and e (recessive ebony body colour). Flies in the next generation that have mutagenize chromosome 3 covered by the balancer, are crossed to flies that have a third-chromosome deletion and the e marker . If the mutagenized third chromosome has a recessive lethal mutation (x) at a site within the deletion, there are viable progeny that do not carry Sb. Then, the recessive lethal is present, balanced against the Sb chromosome in all viable progeny that are not ebony (e/e). (b) When 1/Sb and 2/Sb flies are crossed (1 and 2 are two different lethals), one-third of viable progeny should be wild type (without Sb) if 1 and 2 are in different complementation groups (equivalent to genes( exhibit Sb if 1 and 2 are in the same complementation group.
In the study on which Fig. 2 is based (1), a total of 268 recessive lethals that fall within a segment of chromosome 3 comprising 26 polytene chromosomal bands could be assigned to 25 complementation groups. The numbers were sufficiently great to make it likely that most, if not all, of the genes that have essential functions and therefore can mutate to recessive lethality had been identified. The gene number was probably underestimated because there are certainly genes that can be deleted without lethality. Nevertheless, the agreement between chromosome band number and the apparent number of essential genes (now found in several different experiments) is too close to be without significance.
2. The Use of Heterokaryons
Filamentous fungi, or at least those (eg, Neurospora crassa, Aspergillus nidulans) that have been well studied genetically, are naturally haploid except for the immediate product of sexual fusion, which immediately undergoes meiosis to provide nuclei for haploid spores. In Aspergillus, however, rare vegetative diploids are obtained by selection (2). In Neurospora, where this is not possible, complementation tests are easily carried out with haploid heterokaryons, mixed cultures in which the filamentous hyphae have undergone fusions to bring the two different kinds of haploid nuclei together in the same cytoplasm (3).
For heterokaryon formation, Neurospora crassa strains have to be of the same mating type, and identical with respect to a number of other genes governing vegetative compatibility. These requirements are automatically met if the mutants under test have all been isolated in the same wild-type strain, as is usually the case. Given vegetative compatibility, fusions occur readily as soon as mixed mutant inocula of conidia (asexual spores) begin to germinate. Neurospora filaments (hyphae( are coenocytic. Numerous nuclei are present together in each cytoplasmic compartment, and the nuclear ratio in a heterokaryon is quite variable but is determined at least approximately by the ratio of the two kinds of conidia in the initial mixed inoculum. The nuclear ratio is, in fact, not usually critical. If, as is usually the case, the mutants under test are auxotrophic (ie, have special nutritional requirements) and the mixed inoculation is on a minimal medium on which neither mutant grows by itself, the heterokaryon grows vigorously if the mutants complement one another.
At least in Neurospora, different genes can complement each other perfectly efficiently in heterocaryons even though they are separated in different nuclei. This is not at all surprising. Genes act, after all, by exporting their messenger RNA to the cytoplasm. Reports indicate that complementation in Aspergillus is more efficient in diploids than in heterokaryons, but one suspects that this is because, in this fungus, the different nuclear types are not sufficiently closely intermingled. A large number of Neurospora auxotrophic mutants have been efficiently classified into complementation groups, equivalent to genes, by heterokaryon testing. An example is shown in Fig. 3 (4).

Figure 3. The result of complementation tests by heterokaryon formation on 46 Neurospora mutants that cannot utilize acetate. Nearly all possible pairwise mixtures of mutant conidia were inoculated into a medium containing acetate as the sole carbon source. Growth occurred only if the mutants showed complementation. In the matrix shown, the mutants, designated by their original isolation numbers, are divided into seven different complementation groups. All intergroup combinations showed complementation and grew like wild type. Most or all combinations within a group did not grow. The growth shown by a few pairs of mutants within group 6 is evidence for inter allelic complementation.
Humans, of course, are diploid, but complementation testing by controlled sexual crossing is obviously out of the question. Here again, heterokaryosis may provide an alternative, and cells in culture may serve as surrogates for people. One good example is an analysis of the human xeroderma pigmentosum syndrome, caused by genetic deficiency in DNA repair (5). Cultured cells isolated from different patients were induced to fuse by treating them with heat-inactivated Sendai virus. Complementation in the resulting heterokaryotic cells was assessed by their ability or inability to incorporate a radioactive precursor into their DNA following damage by UV light. The results permitted assigning the different mutations to four different complementation groups, putatively different genes (Fig. 4).

Figure 4. The use of heterokaryosis for complementation testing of cultured cells from human Xeroderma Pigmentosum (XP) patients (5). The syndrome that is excessive sensitivity to uv light is a consequence of failure to repair damaged DNA by excision followed by new synthesis. (a) Cultured fibroblasts from different patients were mixed and induced to undergo fusions. The resulting cells, some now heterokaryotic, were irradiated and cultured in the presence of tritiated thymidine to see whether they responded to the DNA damage by incorporating this radioactive DNA precursor into their nuclei. The original XP mutant cells, or noncomplemented heterokaryons, incorporated no tritium, but certain pairs of mutant cells showed complementation. (b) The complementation matrix with the division of 12 XP mutations into four complementation groups A-D. Here the cells are diploid and homozygous for the different mutations instead of haploid as in the Neurospora example (Fig. 3), but the principle is the same. +, complementation; 0, no complementation; –, test not done.
2.1. In Viruses by Mixed Infection
S. Benzer's very extensive complementation testing of bacteriophage T4 mutants depended on infection of bacterial cells with mixtures of phage particles (6.( Mixed infection is analogous to heterokaryosis because two different whole genomes are present in a somewhat indeterminate ratio, but the difference is that the simple viral genomes are not enclosed in nuclei. The mutants under study (class rII) were distinguished by growing on one Escherichia coli strain (B) but not on another (K). Complementation between pairs of mutants was checked by superimposing rII phage inocula on lawns of K cells, usually with a small admixture of B cells to allow at least some viral propagation. Three results were clearly distinguished: (1) complementation of the two mutants—total clearing of the mixedly infected patch due to lysis of all the cells in that area; (2) no complementation, but some formation of wild-type phage by recombination during mixed growth, shown by scattered spots of lysis; and (3) much less commonly, neither complementation nor recombination—no lysis at all (Fig. 5a). These results were interpreted to mean that (1) mutant sites are in different cistrons (ie, different functional genes); (2) mutants are at different sites in the same cistron; and (3) mutants are at the same site, or one is a deletion overlapping the other. The complementation results split the rII mutants cleanly into two mutually complementing groups or cistrons. Recombination analysis using a set of overlapping deletion mutants showed that these two cistrons correspond to two adjacent, nonoverlapping segments of the phage “chromosome”, which is not actually a chromosome in the cytological sense but a single molecule of double-stranded DNA.
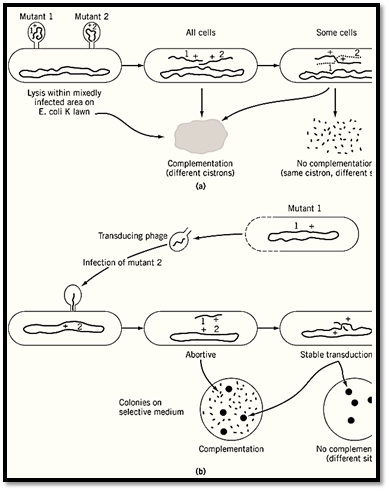
Figure 5. Two examples from bacteria of complementation tests that distinguish between complementation and recombine)a) Testing for complementation between rII mutants of bacteriophage T4. These mutants grow well in Escherichia coli B but not at all in strain K12(l). Mixed infections on a lawn of K cells that have a minor admixture of B cells to permit sojoint growth give total clearing (lysis) if mutants rII-1 and rII-2 are in different genes (cistrons), and lysis only in spots bof recombination, if they are at different sites in the same gene. Based on Ref. 6. (b) Abortive transduction as a criterion complementation in Salmonella typhimurium. Phage P22 grown on histidine-requiring (his-) mutant 1 was used to infect mutant 2. Surviving bacteria were tested for their ability to grow on a histidine-free medium. Rather numerous tiny (abortive colonies showed that a nonreplicating fragment of DNA from mutant 1 complements mutant 2. The absence of such tiny colonies signifies no complementation. In either case, vigorously growing colones arise by recombination between the b: chromosome and the incoming fragment, provided that 1 and 2 are at different sites.
2.2. By Partial Diploids (Merodiploids) in Bacteria
It is not possible to test in bacteria for complementation between whole genomes because, at least in those species that are well-analyzed genetically, there is neither a stable diploid phase nor any sexual process that transfers a whole genome from one cell to another. However, bacteria acquire extra genomic fragments in three general ways: (1) by infection with defective bacteriophages which, “by mistake”, have picked up bacterial DNA (transduction); (2) by joining a fragment of bacterial DNA to the DNA of a plasmid that brings about its own intracellular transfer (conjugation); and (3) by uptake of free DNA (transformation). The first two methods have been important in complementation analysis.
Transduction has been the principal means of classification and mapping of auxotrophic mutants of the bacterium Salmonella typhimurium. Bacteriophage P22 grown on one mutant strain (the donor( is used to infect a second mutant strain, perhaps requiring supplementation with the same nutrient, such as an amino acid. Most of the infected bacteria are lysed, but some survive, are now lysogenic, and harbor the phage in latent form. Some survivors have acquired some fragment or other of the genome of the donor bacterium by infection from defective bacteriophage. The donor DNA is replicated in the recipient cell only if it recombines into the recipient chromosome and replaces the equivalent recipient segment. More commonly the donor DNA persists without replication, and its genes are expressed only in the primary recipient cell and in a few surrounding cells by diffusion of gene products. The appearance of tiny colonies of cells (abortive transductants( that grow transiently without the nutritional supplement, demonstrates complementation between the recipient genome and the donor genomic fragment. Usually a much smaller number of large strongly growing colonies results from chromosomal integration of a donor fragment and recombination between donor and recipient mutational sites to reconstitute a wild-type gene. The presence of large colonies without abortive ones shows that the mutations are at different sites within the same gene or cistron (Fig. 5b). Failure to form any colonies at all means (apart from possible experimental error( that the two mutations are at the same site or that one is a deletion overlapping the other (7).
The plasmid that is used to bring about partial diploidy in E. coli is the transmissible “sex factor” F, a closed-loop DNA molecule that replicates autonomously in the bacterial cell and is occasionally integrated by crossing-over into the much larger loop of the bacterial chromosome. The integrated plasmid DNA undergoes occasional excision, usually precisely, but sometimes carries with it an adjacent segment of bacterial DNA, the nature of which depends on where the plasmid happened to have been inserted. This modified F is called F′ and, like F itself, is transmitted from cell to cell. F′-lac plasmids that carry segments of DNA from the lac operon of the bacterial chromosome, were of great importance in elucidating the complementation relationships of E. coli lac mutants (a topic dealt with separately under cis-dominance).
References
1. J. Gausz, H. Gyurovics, G. Beneze et al. (1981) Genetics 98, 775–789.
2. J. A. Roper (1952). Experientia 8, 14–15.
3. G. W. Beadle and V. L. Coonradt (1944) Genetics 29, 291–308.
4. R. B. Flavell and J. R. S. Fincham (1968) J. Bacteriol. 95, 1056–1062.
5. K. H. Kraemer, H. G. Coon, R. A. Petinga et al. (1975) Proc. Natl. Acad. Sci. USA 72, 59–63.
6. S. Benzer (1959) Proc. Natl. Acad. Sci. USA 45, 1607–1620.
7. P. E. Hartman, Z. Hartman, and D. Serman (1960) J. Gen. Microbiol. 22, 354–358.



|
|
|
|
دراسة يابانية لتقليل مخاطر أمراض المواليد منخفضي الوزن
|
|
|
|
|
|
|
اكتشاف أكبر مرجان في العالم قبالة سواحل جزر سليمان
|
|
|
|
|
|
|
المجمع العلمي ينظّم ندوة حوارية حول مفهوم العولمة الرقمية في بابل
|
|
|