


 النبات
النبات
 الحيوان
الحيوان
 الأحياء المجهرية
الأحياء المجهرية
 علم الأمراض
علم الأمراض
 التقانة الإحيائية
التقانة الإحيائية
 التقنية الحيوية المكروبية
التقنية الحيوية المكروبية
 التقنية الحياتية النانوية
التقنية الحياتية النانوية
 علم الأجنة
علم الأجنة
 الأحياء الجزيئي
الأحياء الجزيئي
 علم وظائف الأعضاء
علم وظائف الأعضاء
 الغدد
الغدد
 المضادات الحيوية
المضادات الحيوية|
Read More
Date: 14-12-2015
Date: 31-3-2021
Date: 6-12-2015
|
Introduction to The Eukaryotic Transcription
KEY CONCEPT
- Chromatin must be opened before RNA polymerase can bind the promoter.
Initiation of transcription on a chromatin template that is already opened requires the enzyme RNA polymerase to bind at the promoter and transcription factors to bind to enhancers. In vitro
transcription on a DNA template requires a different subset of transcription factors than are needed to transcribe a chromatin template (we examine how chromatin is opened in the chapter titled Eukaryotic Transcription Regulation). Any protein that is needed for the initiation of transcription, but that is not itself part of RNA polymerase, is defined as a transcription factor. Many transcription factors act by recognizing cis-acting sites on DNA. Binding to DNA, however, is not the only means of action for a transcription factor.
A factor may recognize another factor, recognize RNA polymerase, or be incorporated into an initiation complex only in the presence of several other proteins. The ultimate test for membership in the transcription apparatus is functional: A protein must be needed for transcription to occur at a specific promoter or set of promoters.
A significant difference between the transcription of eukaryotic and prokaryotic RNAs is that in bacteria transcription takes place on a DNA template, whereas in eukaryotes transcription takes place on a chromatin template. Chromatin changes everything and must be taken into account at every step. The chromatin must be in an open structure, and, even in an open structure, nucleosome octamers must be moved or removed from promoter sequences before transcription factors and RNA polymerase can bind. This can sometimes require transcription from a silent or cryptic promoter either on the same strand or on the antisense strand.
A second major difference is that the bacterial RNA polymerase, with its sigma factor subunit, can read the DNA sequence to find and bind to its promoter. A eukaryotic RNA polymerase cannot read DNA. Initiation at eukaryotic promoters therefore involves a large number of factors that must prebind to a variety of cis-acting elements and other factors already bound to the DNA before the RNA polymerase can bind. These factors are called basal transcription factors. The RNA polymerase then binds to this basal transcription factor–DNA complex. This binding region is defined as the core promoter, the region containing all the binding sites necessary for RNA polymerase to bind and function. RNA polymerase itself binds around the start point of transcription, but does not directly contact the extended upstream region of the promoter. By contrast, bacterial promoters discussed in the chapter titled Prokaryotic Transcription are largely defined in terms of the binding site for RNA polymerase in the immediate vicinity of the start point.
Whereas bacteria have a single RNA polymerase that transcribes all three major classes of genes, transcription in eukaryotic cells is divided into three classes. Each class is transcribed by a different RNA polymerase:
-RNA polymerase I transcribes 18S/28S rRNA.
-RNA polymerase II transcribes mRNA and some small RNAs.
-RNA polymerase III transcribes tRNA, 5S ribosomal RNA, and also some other small RNAs.
This is the current picture of the major classes of genes. As we will see in the chapter titled Regulatory RNA, recent discoveries by whole genome tiling arrays and deep sequencing of cellular RNA have uncovered a new world of antisense transcripts, intergenic transcripts, and heterochromatin transcripts. Virtually the entire genome is transcribed from both strands. Not much is currently known about the promoters for these classes or their function and regulation, but it is known that many (possibly most) of these transcripts are produced by RNA polymerase II.
Basal transcription factors are needed for initiation, but most are not required subsequently. For the three eukaryotic RNA polymerases, the transcription factors, rather than the RNA polymerases themselves, are responsible for recognizing the promoter DNA sequence. For all eukaryotic RNA polymerases, the basal transcription factors create a structure at the promoter to provide the target that is recognized by the RNA polymerase. For RNA polymerases I and III, these factors are relatively simple, but for RNA polymerase II they form a sizeable group. The basal factors join with RNA polymerase II to form a complex surrounding the start point, and they determine the site of initiation. The basal factors together with RNA polymerase constitute the basal transcription apparatus.
The promoters for RNA polymerases I and II are (mostly) upstream of the start point, but a large number of promoters for RNA polymerase III lie downstream (within the transcription unit) of the start point. Each promoter contains characteristic sets of short conserved sequences that are recognized by the appropriate class of basal transcription factors. RNA polymerases I and III each recognize a relatively restricted set of promoters, and thus rely upon a small number of accessory factors.
Promoters utilized by RNA polymerase II show much more variation in sequence and have a modular organization. All RNA polymerase II promoters have sequence elements close to the start point of transcription that are bound by the basal apparatus and the polymerase to establish the site of initiation. Other sequences farther upstream or downstream, called enhancer sequences, determine whether the promoter is expressed, and, if expressed, whether this occurs in all cell types or is cell type specific.
The enhancer is a second type of site involved in transcription and is identified by sequences that stimulate initiation. Enhancer elements are often targets for tissue-specific or temporal regulation. Some enhancers bind transcription factors that function by short-range interactions and are located near the promoter, whereas others can be located thousands of base pairs away.
FIGURE .1 illustrates the general properties of promoters and enhancers. A regulatory site that binds more negative regulators than positive regulators to control transcription is called a silencer. As can be seen in Figure .1, promoters and enhancers are sequences that bind a variety of proteins that control transcription, and in that regard are actually quite similar to each other.
Enhancers, like promoters, can also bind RNA polymerase and initiate transcription of an RNA called eRNA (enhancer RNA) as discussed in the chapter called Regulatory RNA. These eRNAs may promote enhancer/promoter interactions by DNA looping, often through intermediates called coactivators. The components of an enhancer or a silencer resemble those of the promoter in that they consist of a variety of modular elements that can bind positive regulators or negative regulators in a closely packed array. Enhancers do not need to be near the promoter. They can be upstream, inside a gene, or beyond the end of a gene, and their orientation relative to the gene does not matter.
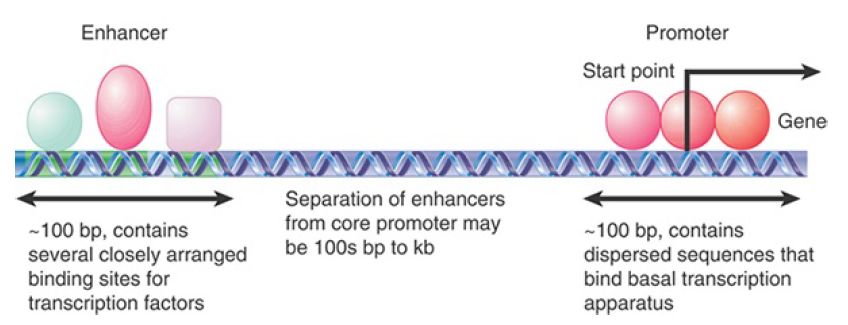
FIGURE .1 A typical gene transcribed by RNA polymerase II has a promoter that extends upstream from the site where transcription is initiated. The promoter contains several short-sequence (~10 bp) elements that bind transcription factors, dispersed over ~100 bp.
An enhancer containing a more closely packed array of elements that also bind transcription factors may be located several hundred base pairs to several kilobases distant. (DNA may be coiled or otherwise rearranged so that transcription factors at the promoter and at the enhancer interact to form a large protein complex.)
Promoters that are constitutively expressed and needed in all cells (their genes are sometimes called housekeeping genes) have upstream sequence elements that are recognized by ubiquitous activators. No one element/factor combination is an essential component of the promoter, which suggests that initiation by RNA polymerase II may be regulated in many different ways. Promoters that are expressed only in certain times or places have sequence elements that require activators that are available only at those times or places.
Because chromatin is a general negative regulator, eukaryotic transcription is most often under positive regulation: A transcription factor is provided under tissue-specific control to activate a
promoter or set of promoters that contain a common target sequence. This is a multistep process that first involves opening the chromatin and binding the basal transcription factors, and then binding the polymerase. Regulation by specific repression of a target promoter is less common.
A eukaryotic transcription unit generally contains a single gene, and termination typically occurs beyond the end of the coding region. Termination lacks the regulatory importance that applies in prokaryotic systems. RNA polymerases I and III terminate at discrete sequences in defined reactions, but the mode of termination by RNA polymerase II is not clear. The significant event in generating the 3′ end of an mRNA, however, is not the termination event itself, but rather a cleavage reaction in the primary transcript .



|
|
|
|
دراسة: إجراء واحد لتقليل المخاطر الجينية للوفاة المبكرة
|
|
|
|
|
|
|
"الملح والماء" يمهدان الطريق لأجهزة كمبيوتر تحاكي الدماغ البشري
|
|
|
|
|
|
جمعيّة العميد وقسم الشؤون الفكريّة تدعوان الباحثين للمشاركة في الملتقى العلمي الوطني الأوّل
|
|
|
|
الأمين العام المساعد لجامعة الدول العربية السابق: جناح جمعية العميد في معرض تونس ثمين بإصداراته
|
|
|
|
المجمع العلمي يستأنف فعاليات محفل منابر النور في واسط
|
|
|
|
برعاية العتبة العباسيّة المقدّسة فرقة العبّاس (عليه السلام) تُقيم معرضًا يوثّق انتصاراتها في قرية البشير بمحافظة كركوك
|