


 النبات
النبات
 الحيوان
الحيوان
 الأحياء المجهرية
الأحياء المجهرية
 علم الأمراض
علم الأمراض
 التقانة الإحيائية
التقانة الإحيائية
 التقنية الحيوية المكروبية
التقنية الحيوية المكروبية
 التقنية الحياتية النانوية
التقنية الحياتية النانوية
 علم الأجنة
علم الأجنة
 الأحياء الجزيئي
الأحياء الجزيئي
 علم وظائف الأعضاء
علم وظائف الأعضاء
 الغدد
الغدد
 المضادات الحيوية
المضادات الحيوية|
Read More
Date: 29-12-2015
Date: 1-12-2015
Date: 7-6-2021
|
Nonhomologous End Joining Also Repairs Double-Strand Breaks
KEY CONCEPTS
- Repair of double-strand breaks when homologous sequence is not available occurs through a
nonhomologous end joining (NHEJ) reaction.
- The NHEJ pathway can ligate blunt ends of duplex DNA.
- Mutations in double-strand break repair pathways cause human diseases.
Repair of DSBs by homologous recombination ensures that no genetic information is lost from a broken DNA end. In many cases, though, a sister chromatid or homologous chromosome is not easily available to use as a template for repair. In addition, some DSBs are specifically repaired using error-prone mechanisms as an intermediate in the recombination of immunoglobulin genes . In these cases, the mechanism used to repair these breaks is called nonhomologous end joining (NHEJ) and consists of ligating the ends together.
The steps involved in NHEJ are summarized in FIGURE 1. The same enzyme complex undertakes the process in both NHEJ and immune recombination. The first stage is recognition of the broken ends by a heterodimer consisting of the proteins Ku70 and Ku80.
After the DNA ends are bound by the Ku complex, the MRN complex (or MRX complex in yeast) assists in bringing the broken DNA ends together by acting as a bridge between the two molecules. The MRN complex consists of Mre11, Rad50, and Nbs1 (Xrs2 in yeast). Another key component is the DNA-dependent protein kinase (DNA-PKcs ), which is activated by DNA to phosphorylate protein targets. One of these targets is the protein Artemis, which in its activated form has both exonuclease and endonuclease activities and can trim overhanging ends and cleave the hairpins generated by recombination of immunoglobulin genes.
The DNA polymerase activity that fills in any remaining singlestranded protrusions is not known. Frequently during the NHEJ process, mutations are generated through nucleotide deletion and insertion that occurs during the processing steps prior to ligation.
The actual joining of the double-stranded ends is performed by DNA ligase IV, which functions in conjunction with the protein XRCC4. Mutations in any of these components may render eukaryotic cells more sensitive to radiation. Some of the genes for these proteins are mutated in patients who have diseases due to deficiencies in DNA repair.
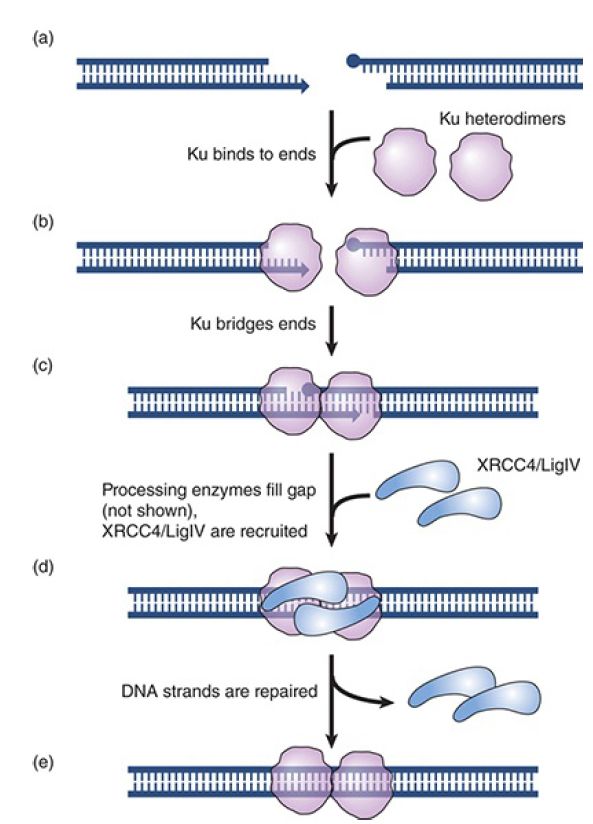
FIGURE 1. Nonhomologous end joining. The blue dot on one of the two double-strand break ends signifies a nonligatable end (a). The double-strand break ends are bound by the Ku heterodimer (b). The Ku–DNA complexes are juxtaposed (c) to bridge the ends, and the gap is filled in by processing enzymes and Pol lambda or Pol mu. The ends are ligated by the specialized DNA ligase LigIV with its partner XRCC4 (d) to repair the double-strand break (e).
The Ku heterodimer is the sensor that detects DNA damage by binding to the broken ends. Ku can bring broken ends together by binding two DNA molecules. The crystal structure in FIGURE 2 shows why it binds only to ends: The bulk of the protein extends for about two turns along one face of DNA (visible in the lower panel), but a narrow bridge between the subunits, located in the center of the structure, completely encircles DNA. This means that the
heterodimer needs to slip onto a free end.
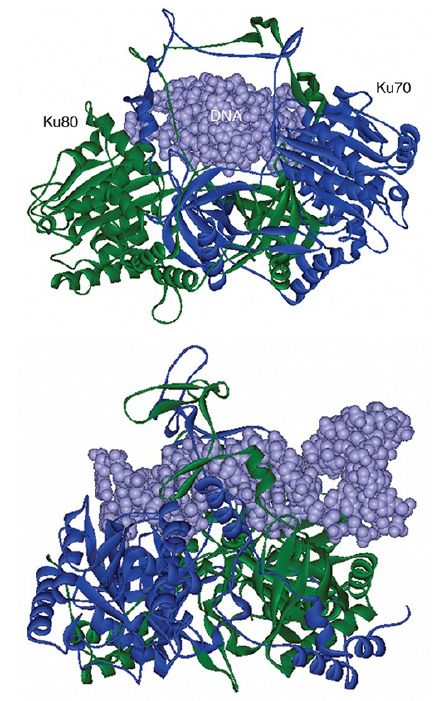
FIGURE 2.The Ku70–Ku80 heterodimer binds along two turns of the DNA double helix and surrounds the helix at the center of the binding site.
Structures from Protein Data Bank 1JEY. J. R. Walker, R. A. Corpina, and J. Goldberg, Nature 412 (2001): 607–614.
All of the repair pathways we have discussed are conserved in mammals, yeast, and bacteria. Deficiency in DNA repair causes several human diseases. The inability to repair DSBs in DNA is particularly severe and leads to chromosomal instability. The instability is revealed by chromosomal aberrations, which are associated with an increased rate of mutation, which, in turn, leads to an increased susceptibility to cancer in patients with the disease.
The basic cause can be mutation in pathways that control DNA repair or in the genes that encode enzymes of the repair complexes. The phenotypes can be very similar, as in the case of
ataxia telangiectasia (AT), which is caused by failure of a cell cycle checkpoint pathway, and Nijmegen breakage syndrome (NBS), which is caused by a mutation of a repair enzyme.
Nijmegen breakage syndrome results from mutations in a gene encoding a protein (variously called Nibrin, p95, or NBS1) that is a component of the Mre11/Rad50/Nbs1 (MRN) repair complex. When human cells are irradiated with agents that induce DSBs, many factors accumulate at the sites of damage, including the components of the MRN complex. After irradiation, the kinase ATM (encoded by the AT gene) phosphorylates NBS1; this activates the complex, which localizes to sites of DNA damage. Subsequent steps involve triggering a checkpoint (a mechanism that prevents the cell cycle from proceeding until the damage is repaired) and recruiting other proteins that are required to repair the damage.
Patients deficient in either ATM or NBS1 are immunodeficient, sensitive to ionizing radiation, and predisposed to develop cancer, especially lymphoid cancers.
The recessive human disorder Bloom syndrome is caused by mutations in a helicase gene (called BLM) that is homologous to recQ of E. coli. The mutation results in an increased frequency of chromosomal breaks and sister chromatid exchanges. BLM associates with other repair proteins as part of a large complex. One of the proteins with which it interacts is hMLH1, a mismatchrepair protein that is the human homolog of bacterial MutL. The yeast homologs of these two proteins, Sgs1 and Mlh1, also associate, identifying these genes as parts of a well-conserved repair pathway and illustrating that there is crosstalk between different repair pathways.



|
|
|
|
تفوقت في الاختبار على الجميع.. فاكهة "خارقة" في عالم التغذية
|
|
|
|
|
|
|
أمين عام أوبك: النفط الخام والغاز الطبيعي "هبة من الله"
|
|
|
|
|
|
|
قسم شؤون المعارف ينظم دورة عن آليات عمل الفهارس الفنية للموسوعات والكتب لملاكاته
|
|
|