


 النبات
النبات
 الحيوان
الحيوان
 الأحياء المجهرية
الأحياء المجهرية
 علم الأمراض
علم الأمراض
 التقانة الإحيائية
التقانة الإحيائية
 التقنية الحيوية المكروبية
التقنية الحيوية المكروبية
 التقنية الحياتية النانوية
التقنية الحياتية النانوية
 علم الأجنة
علم الأجنة
 الأحياء الجزيئي
الأحياء الجزيئي
 علم وظائف الأعضاء
علم وظائف الأعضاء
 الغدد
الغدد
 المضادات الحيوية
المضادات الحيوية|
Read More
Date: 2-11-2020
Date: 1-11-2020
Date: 21-4-2016
|
Each Eukaryotic Chromosome Contains Many Replicons
KEY CONCEPTS
- A chromosome is divided into many replicons.
- The progression into S phase is tightly controlled.
- Eukaryotic replicons are 40 to 100 kilobases (kb) in length.
- Individual replicons are activated at characteristic times during S phase.
- Regional activation patterns suggest that replicons near one another are activated at the same time.
In eukaryotic cells, the replication of DNA is confined to the second part of the cell cycle, called S phase, which follows the G1 phase (see the chapter titled Replication Is Connected to the Cell Cycle).
The eukaryotic cell cycle is composed of alternating rounds of growth followed by DNA replication and cell division. After the cell divides into two daughter cells, each must grow back to approximately the size of the original mother cell before cell division can occur again. The G1 phase of the cell cycle is primarily concerned with growth (although G1 is an abbreviation for first gap because the early cytologists could not see any activity). In G1, everything except DNA begins to be doubled: RNA, protein, lipids, and carbohydrate. The progression from G1 into S is very tightly regulated and controlled by a checkpoint. For a cell to be allowed to progress into S phase, there must be a certain minimum amount of growth, which is biochemically measured. In addition, there must not be any damage to the DNA. Damaged DNA or too little growth prevents the cell from progressing into S phase. When S phase is completed, G2 phase commences. There is no control point and no sharp demarcation.
Replication of the large amount of DNA contained in eukaryotic chromatin is accomplished by dividing it into many individual replicons, as shown in FIGURE 1. Only some of these replicons are engaged in replication at any point in S phase.
Presumably, each replicon is activated at a specific time during S phase, although the evidence on this issue is not decisive. Note that a crucial difference between replication in bacteria and replication in eukaryotes is that in bacteria replication is occurring on DNA, whereas in eukaryotes replication is occurring on chromatin and nucleosomes play a role, so their presence must be taken into account. This is discussed in the chapter titled Chromatin.
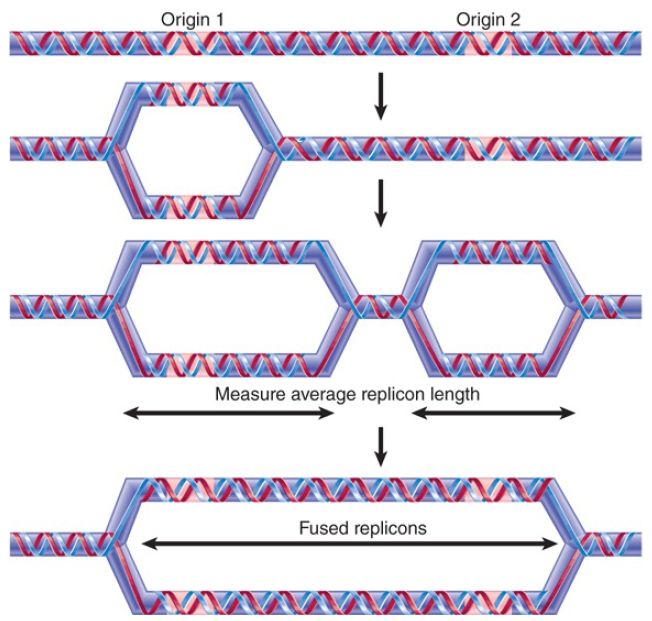
FIGURE 1. A eukaryotic chromosome contains multiple origins of replication that ultimately merge during replication.
The start of S phase is signaled by the activation of the first replicons. Over the next few hours, initiation events occur at other replicons in an ordered manner. Chromosomal replicons usually display bidirectional replication.
Individual replicons in eukaryotic genomes are relatively small, typically approximately 40 kb in yeast or flies and approximately 100 kb in animal cells. They can, however, vary more than 10-fold in length within a genome. The rate of replication is approximately 2,000 bp/min, which is much slower than the 50,000 bp/min of bacterial replication fork movement, presumably because the chromosome is assembled into chromatin, not naked DNA.
From the speed of replication, it is evident that a mammalian genome could be replicated in approximately 1 hour if all replicons functioned simultaneously. S phase actually lasts for more than 6 hours in a typical somatic cell, though, which implies that no more than 15% of the replicons are likely to be active at any given moment. There are some exceptional cases, such as the early embryonic divisions of Drosophila embryos, and other organisms that do not have the leisure of placental development, for which the duration of S phase is compressed by the simultaneous functioning of a large number of replicons.
How are origins selected for initiation at different times during S phase? In Saccharomyces cerevisiae, the default appears to be for replicons to replicate early, but cis-acting sequences can cause origins linked to them to replicate at a later time. In other organisms, there is a general hierarchy to the order of replication. Replicons near active genes are replicated earliest and replicons in heterochromatin replicate last.
Available evidence suggests that most chromosomal replicons do not have a termination region like that of bacteria at which the replication forks cease movement and (presumably) dissociate
from the DNA. It seems more likely that a replication fork continues from its origin until it meets a fork proceeding toward it from the adjacent replicon. Recall the discussion about the potential topological problem of joining the newly synthesized DNA at the junction of the replication forks.
The propensity of replicons located in the same vicinity to be active at the same time could be explained by “regional” controls, in which groups of replicons are initiated more or less coordinately, as opposed to a mechanism in which individual replicons are activated one by one in dispersed areas of the genome. Two structural features suggest the possibility of large-scale organization. Quite large regions of the chromosome can be characterized as “early replicating” or “late replicating,” implying that there is little interspersion of replicons that fire at early or late times.
Visualization of replicating forks by labeling with DNA precursors identifies 100 to 300 “foci” instead of uniform staining; each focus shown in FIGURE 2 probably contains greater than 300
replication forks. The foci could represent fixed structures through which replicating DNA must move.

FIGURE 2. Replication forks are organized into foci in the nucleus. Cells were labeled with BrdU. The left panel was stained with propidium iodide to identify bulk DNA. The right panel was stained using an antibody to BrdU to identify replicating DNA.
Photos courtesy of Anthony D. Mills and Ron Laskey, Hutchinson/MRC Research Center, University of Cambridge.



|
|
|
|
تفوقت في الاختبار على الجميع.. فاكهة "خارقة" في عالم التغذية
|
|
|
|
|
|
|
أمين عام أوبك: النفط الخام والغاز الطبيعي "هبة من الله"
|
|
|
|
|
|
|
قسم شؤون المعارف ينظم دورة عن آليات عمل الفهارس الفنية للموسوعات والكتب لملاكاته
|
|
|