

النبات

مواضيع عامة في علم النبات

الجذور - السيقان - الأوراق

النباتات الوعائية واللاوعائية

البذور (مغطاة البذور - عاريات البذور)

الطحالب

النباتات الطبية


الحيوان

مواضيع عامة في علم الحيوان

علم التشريح

التنوع الإحيائي

البايلوجيا الخلوية


الأحياء المجهرية

البكتيريا

الفطريات

الطفيليات

الفايروسات


علم الأمراض

الاورام

الامراض الوراثية

الامراض المناعية

الامراض المدارية

اضطرابات الدورة الدموية

مواضيع عامة في علم الامراض

الحشرات


التقانة الإحيائية

مواضيع عامة في التقانة الإحيائية


التقنية الحيوية المكروبية

التقنية الحيوية والميكروبات

الفعاليات الحيوية

وراثة الاحياء المجهرية

تصنيف الاحياء المجهرية

الاحياء المجهرية في الطبيعة

أيض الاجهاد

التقنية الحيوية والبيئة

التقنية الحيوية والطب

التقنية الحيوية والزراعة

التقنية الحيوية والصناعة

التقنية الحيوية والطاقة

البحار والطحالب الصغيرة

عزل البروتين

هندسة الجينات


التقنية الحياتية النانوية

مفاهيم التقنية الحيوية النانوية

التراكيب النانوية والمجاهر المستخدمة في رؤيتها

تصنيع وتخليق المواد النانوية

تطبيقات التقنية النانوية والحيوية النانوية

الرقائق والمتحسسات الحيوية

المصفوفات المجهرية وحاسوب الدنا

اللقاحات

البيئة والتلوث


علم الأجنة

اعضاء التكاثر وتشكل الاعراس

الاخصاب

التشطر

العصيبة وتشكل الجسيدات

تشكل اللواحق الجنينية

تكون المعيدة وظهور الطبقات الجنينية

مقدمة لعلم الاجنة


الأحياء الجزيئي

مواضيع عامة في الاحياء الجزيئي


علم وظائف الأعضاء


الغدد

مواضيع عامة في الغدد

الغدد الصم و هرموناتها

الجسم تحت السريري

الغدة النخامية

الغدة الكظرية

الغدة التناسلية

الغدة الدرقية والجار الدرقية

الغدة البنكرياسية

الغدة الصنوبرية

مواضيع عامة في علم وظائف الاعضاء

الخلية الحيوانية

الجهاز العصبي

أعضاء الحس

الجهاز العضلي

السوائل الجسمية

الجهاز الدوري والليمف

الجهاز التنفسي

الجهاز الهضمي

الجهاز البولي


المضادات الميكروبية

مواضيع عامة في المضادات الميكروبية

مضادات البكتيريا

مضادات الفطريات

مضادات الطفيليات

مضادات الفايروسات

علم الخلية

الوراثة

الأحياء العامة

المناعة

التحليلات المرضية

الكيمياء الحيوية

مواضيع متنوعة أخرى

الانزيمات
The Spliceosome Assembly Pathway
المؤلف:
JOCELYN E. KREBS, ELLIOTT S. GOLDSTEIN and STEPHEN T. KILPATRICK
المصدر:
LEWIN’S GENES XII
الجزء والصفحة:
12-5-2021
3355
The Spliceosome Assembly Pathway
KEY CONCEPTS
- The commitment complex progresses to prespliceosome (the A complex) in the presence of ATP.
- Binding of U5 and U4/U6 snRNPs converts the A complex to the mature spliceosome (the B1 complex).
- The B1 complex is next converted to the B2 complex, in which U1 snRNP is released to allow U6 snRNA to interact with the 5′ splice site.
- When U4 dissociates from U6 snRNP, U6 snRNA can pair with U2 snRNA to form the catalytic active site.
- Both transesterification reactions take place in the activated spliceosome (the C complex).
- The splicing reaction is reversible at all steps.
Following formation of the E complex, the other snRNPs and factors involved in splicing associate with the complex in a defined order. FIGURE 1 shows the components of the complexes that can be identified as the reaction proceeds.

FIGURE 1 The splicing reaction proceeds through discrete stages in which spliceosome formation involves the interaction of components that recognize the consensus sequences.
In the first ATP-dependent step, U2 snRNP joins U1 snRNP on the pre-mRNA by binding to the branch point sequence, which also involves base pairing between the sequence in U2 snRNA and the branch point sequence. This results in the conversion of the E complex to the prespliceosome commonly known as the A complex, and this step requires ATP hydrolysis.
The B1 complex is formed when a trimer containing the U5 and U4/U6 snRNPs binds to the A complex. This complex is regarded as a spliceosome because it contains the components needed for the splicing reaction. It is converted to the B2 complex after U1 is released. The dissociation of U1 is necessary to allow other components to come into juxtaposition with the 5′ splice site, most notably U6 snRNA.
The catalytic reaction is triggered by the release of U4, which also takes place during the transition from the B1 to B2 complex. The role of U4 snRNA may be to sequester U6 snRNA until it is needed.
FIGURE 2 shows the changes that occur in the base-pairing interactions between snRNAs during splicing. In the U6/U4 snRNP, a continuous length of 26 bases of U6 is paired with two separated regions of U4. When U4 dissociates, the region in U6 that is released becomes free to take up another structure. The first part of it pairs with U2; the second part forms an intramolecular hairpin. The interaction between U4 and U6 is mutually incompatible with the interaction between U2 and U6, so the release of U4 controls the ability of the spliceosome to proceed to the activated state.

FIGURE 2. U6/U4 pairing is incompatible with U6/U2 pairing. When U6 joins the spliceosome it is paired with U4. Release of U4 allows a conformational change in U6; one part of the released sequence forms a hairpin and the other part pairs with U2. An adjacent region of U2 is already paired with the branch site, which brings U6 into juxtaposition with the branch. Note that the substrate RNA is reversed from the usual orientation and is shown 3′ to 5′.
For clarity, Figure 2 shows the RNA substrate in extended form, but the 5′ splice site is actually close to the U6 sequence immediately on the 5′ side of the stretch bound to U2. This sequence in U6 snRNA pairs with sequences in the intron just downstream of the conserved GU at the 5′ splice site (mutations that enhance such pairing improve the efficiency of splicing).
Thus, several pairing reactions between snRNAs and the substrate RNA occur in the course of splicing. They are summarized in FIGURE 3. The snRNPs have sequences that pair with the pre-mRNA substrate and with one another. They also have singlestranded regions in loops that are in close proximity to sequences in the substrate and that play an important role, as judged by the ability of mutations in the loops to block splicing.
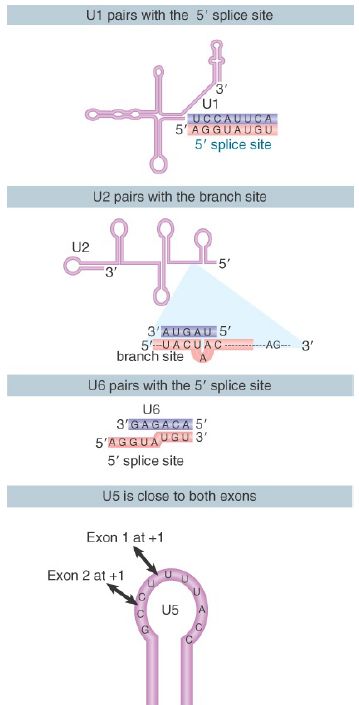
FIGURE 3. Splicing utilizes a series of base-pairing reactions between snRNAs and splice sites.
The base pairings between U2 and the branch point and between U2 and U6 create a structure that resembles the active center of group II self-splicing introns . This suggests the possibility that the catalytic component could comprise an RNA structure generated by the U2–U6 interaction. U6 is paired with the 5′ splice site, and cross-linking experiments show that a loop in U5 snRNA is immediately adjacent to the first base positions in both exons.
Although the available evidence points to an RNA-based catalysis mechanism within the spliceosome, contribution(s) by proteins cannot be ruled out. One candidate protein is Prp8, a large scaffold protein that directly contacts both the 5′ and 3′ splice sites within the spliceosome.
Both transesterification reactions take place in the activated spliceosome (the C complex) after a series of RNA arrangements is completed. The formation of the lariat at the branch site is responsible for determining the use of the 3′ splice site, because the 3′ consensus sequence nearest to the 3′ side of the branch becomes the target for the second transesterification.
The important conclusion suggested by these results is that the snRNA components of the splicing apparatus interact both among themselves and with the substrate pre-mRNA by means of basepairing interactions, and these interactions allow for changes in structure that may bring reacting groups into apposition and may even create catalytic centers.
Although (like ribosomes) the spliceosome is likely a large RNA machine, many protein factors are essential for the machine to run. Extensive mutational analyses undertaken in yeast identified both the RNA and protein components (known as PRP mutants for premRNA processing). Several of the products of these genes have motifs that identify them as a family of ATP-dependent RNA helicases, which are crucial for a series of ATP-dependent RNA rearrangements in the spliceosome.
Prp5 is critical for U2 binding to the branch point during the transition from the E to the A complex; Brr2 facilitates U1 and U4 release during the transition from the B1 to B2 complex; Prp2 is responsible for the activation of the spliceosome during the conversion of the B2 complex to the C complex; and Prp22 helps the release of the mature mRNA from the spliceosome. In addition, a number of RNA helicases play roles in recycling of snRNPs for the next round of spliceosome assembly.
These findings explain why ATP hydrolysis is required from various steps of the splicing reaction, although the actual transesterification reactions do not require ATP. Despite the fact that a sequential series of RNA arrangements takes place in the spliceosome, it is remarkable that the process seems to be reversible after both the first and second transesterification reactions.
 الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
 اخر الاخبار
اخر الاخبار
اخبار العتبة العباسية المقدسة

الآخبار الصحية















 قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة
قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة "المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة
"المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة (نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)
(نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)


















