


 النبات
النبات
 الحيوان
الحيوان
 الأحياء المجهرية
الأحياء المجهرية
 علم الأمراض
علم الأمراض
 التقانة الإحيائية
التقانة الإحيائية
 التقنية الحيوية المكروبية
التقنية الحيوية المكروبية
 التقنية الحياتية النانوية
التقنية الحياتية النانوية
 علم الأجنة
علم الأجنة
 الأحياء الجزيئي
الأحياء الجزيئي
 علم وظائف الأعضاء
علم وظائف الأعضاء
 الغدد
الغدد
 المضادات الحيوية
المضادات الحيوية|
Read More
Date: 3-12-2015
Date: 16-6-2021
Date: 6-4-2021
|
Aminoacyl-tRNA Synthetases Fall into Two Classes
KEY CONCEPT
- Aminoacyl-tRNA synthetases are divided into class I and class II families based on mutually exclusive sets of sequence motifs and structural domains.
In spite of their common function, synthetases are a very diverse group of enzymes. They are divisible into two classes. Class I tRNA synthetases are primarily monomeric and feature structurally similar active-site Rossmann-fold domains at or near their Ntermini.
The Rossmann fold consists of a five- or six-stranded parallel β-sheet with connecting helices. This domain is homologous to the active site domain of dehydrogenases and is responsible for binding the ATP, the amino acid, and the 3′ terminus of tRNA. All class I tRNA synthetases contain an “acceptor-binding” domain that is inserted into the Rossmann fold at a common location, which also binds the single-stranded acceptor end of the tRNA, and which contains an editing active site in some of the enzymes . The C-terminal domains of class I synthetases bind the inner corner of the L-shaped tRNA and the anticodon arm and also function to discriminate among tRNAs. Two short common sequence motifs involved in ATP binding are found in the active-site Rossmann fold. Aside from some limited homology among a few of the enzymes, there are no significant structural or sequence similarities among class I enzymes outside of the Rossmann fold.
Class II tRNA synthetases are similarly diverse. Their quaternary structures are generally dimeric but in some cases form homotetramers or α2 β2 heterotetramers. Like class I enzymes, class II tRNA synthetases also possess a structurally conserved active site domain—in this case a mixed α/β domain dissimilar to the Rossmann fold. The active sites of class II tRNA synthetases are located toward the C-terminal end of the polypeptides. Three short sequence motifs in the active site domain are conserved in this class; one of these motifs functions in multimerization, whereas the other two have catalytic roles.
The tRNA synthetases are grouped into 23 phylogenetically distinct families. Eleven of these families fall into class I; the remaining 12 are class II enzymes (TABLE 1). Interestingly, two distinct types of LysRS enzymes fall into separate classes. Two noncanonical tRNA synthetase families with limited phylogenetic scope have also recently been discovered. These enzymes are the class II pyrrolysyl-tRNA synthetase (PylRS) (discussed in the section earlier in this chapter titled Novel Amino Acids Can Be Inserted at Certain Stop Codons) and the class II phosphoseryltRNA synthetase (SepRS). SepRS is restricted to methanogens (a subclass of archaea) and the closely related Archaeoglobus fulgidus. It attaches phosphoserine (Sep) onto tRNACys acceptors to produce a misacylated Sep-tRNACys type. All organisms possessing SepRS also possess a pyridoxal phosphate-dependent companion enzyme, SepCysS, which converts Sep-tRNACys to Cys-tRNACys . The sulfur donor used by SepCysS in vivo is unknown. Interestingly, some methanogens possess both the SepRS/SepCysS two-step pathway and, in parallel, the canonical CysRS enzyme. Recently, phosphoserine was cotranslationally inserted (in response to the UAG stop codon) into several recombinant proteins made in E. coli by introducing the SepRS enzyme together with an engineered version of elongation factor Tu. This new system holds enormous promise for the study of selectively phosphoserylated proteins such as those involved in signal transduction in mammalian cells.
TABLE 1 Separation of tRNA synthetases into two classes possessing mutually exclusive sets of sequence motifs and activesite structural domains. The quaternary structure of the enzyme is
noted. Multiple designations indicate that the quaternary structure differs in different organisms. The quaternary structure of PylRS has not been clearly established.
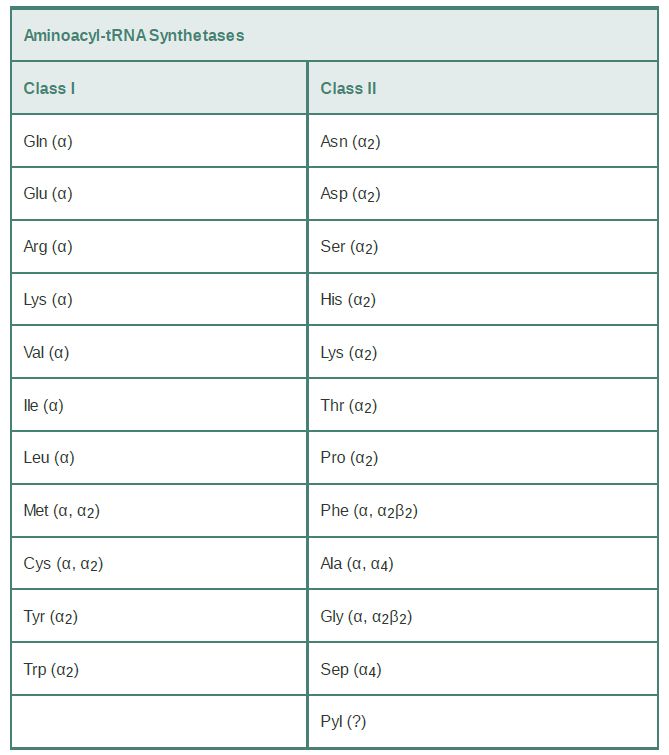
Although there are 23 phylogenetically distinct tRNA synthetase families, most organisms possess only 18 of the enzymes.
Typically missing from the repertoire are GlnRS and asparaginyltRNA synthetase (AsnRS). To synthesize Gln-tRNAGln and AsntRNAAsn , these organisms possess distinct glutamyl-tRNA synthetase (GluRS) and aspartyl-tRNA synthetase (AspRS) enzymes that are nondiscriminating (ND). GluRSND synthesizes both Glu-tRNAGlu as well as misacylated Glu-tRNAGlu ; AspRSND synthesizes both Asp-tRNAAsp and misacylated Asp-tRNAAsn . The misacylated tRNAs are then converted to Gln-tRNAGln and AsntRNAAsn by the action of a tRNA-dependent amidotransferase (AdT). AdTs are remarkable multimeric enzymes possessing three distinct activities (FIGURE 1). They first generate ammonia in one active site by deamidation of a nitrogen donor such as glutamine or asparagine. The ammonia is then shuttled through an intramolecular tunnel in the enzyme to emerge in a second site that binds the 3′ end of the misacylated tRNA. In the second active site, a kinase activity γ-phosphorylates the side-chain amino acid carboxylate of Glu-tRNAGlu or Asp-tRNAAsn . Finally, the ammonia reacts to displace phosphate, forming Gln-tRNAGln or Asn-tRNAAsn . Distinct AdT families that function on both misacylated tRNAs or that are restricted to Gln-tRNAGln formation only also exist.
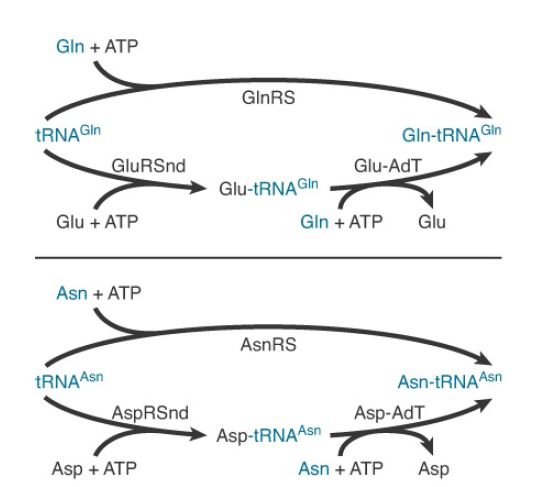
FIGURE 23.13 Mechanisms for the synthesis of Gln-tRNAGln and Asn-tRNAAsn . The top route in each case indicates the one-step pathway catalyzed by the conventional tRNA synthetase. The bottom, two-step pathways are found in most organisms. They consist of a nondiscriminating tRNA synthetase followed by the action of a tRNA-dependent amidotransferase (AdT).
Class I and class II synthetases are functionally differentiated in a number of ways. First, class I enzymes aminoacylate tRNA at the 2′–OH position of A76, whereas class II enzymes generally aminoacylate tRNA on the 3′–OH. The position of initial aminoacylation is related to the binding orientation of the tRNA on the enzyme. Class I synthetases bind tRNA on the minor groove side of the acceptor stem and require that the single-stranded 3′ terminus form a hairpin structure for proper juxtaposition with the amino acid and ATP in the active site (Figure 2). Class II synthetases instead bind the major groove side of the tRNA acceptor stem and do not require hairpinning of the tRNA 3′ end into the active site. A mechanistic distinction also exists: The reaction rates of class I synthetases are limited by release of aminoacylated tRNA product, whereas class II synthetases are limited by earlier chemical steps and/or physical rearrangements in the active sites.
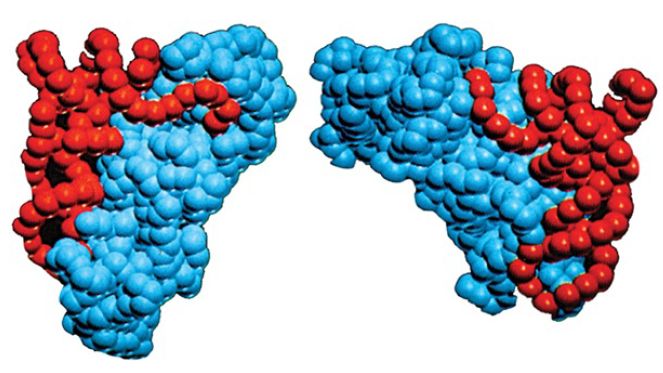
FIGURE 2. Crystal structures show that class I and class II aminoacyl-tRNA synthetases bind the opposite faces of their tRNA substrates. The tRNA is shown in red and the protein in blue. Photo courtesy of Dino Moras, Institute of Genetics and Molecular and Cellular Biology.



|
|
|
|
تفوقت في الاختبار على الجميع.. فاكهة "خارقة" في عالم التغذية
|
|
|
|
|
|
|
أمين عام أوبك: النفط الخام والغاز الطبيعي "هبة من الله"
|
|
|
|
|
|
|
المجمع العلمي ينظّم ندوة حوارية حول مفهوم العولمة الرقمية في بابل
|
|
|