


 النبات
النبات
 الحيوان
الحيوان
 الأحياء المجهرية
الأحياء المجهرية
 علم الأمراض
علم الأمراض
 التقانة الإحيائية
التقانة الإحيائية
 التقنية الحيوية المكروبية
التقنية الحيوية المكروبية
 التقنية الحياتية النانوية
التقنية الحياتية النانوية
 علم الأجنة
علم الأجنة
 الأحياء الجزيئي
الأحياء الجزيئي
 علم وظائف الأعضاء
علم وظائف الأعضاء
 الغدد
الغدد
 المضادات الحيوية
المضادات الحيوية|
Read More
Date: 15-6-2021
Date: 8-3-2021
Date: 1-4-2021
|
Lesion Bypass Requires Polymerase Replacement
KEY CONCEPTS
- A replication fork stalls when it arrives at damaged DNA.
- The replication complex must be replaced by a specialized DNA polymerase for lesion bypass.
-After the damage has been repaired, the primosome is required to reinitiate replication by reinserting the replication complex.
Damage to chromosomes that is not repaired before replication can be catastrophic and lethal. When the replication complex encounters damaged and modified bases such that it cannot place a complementary base opposite it, the polymerase stops and the replication fork may collapse. A cell has two options to avoid death: recombination or lesion bypass. On the leading strand in E. coli, replication can bypass a thymine dimer and can, with the DnaG primase, reinitiate forward DNA synthesis downstream. This leaves a gap behind the fork, which can be repaired by recombination, described as follows.
In addition, bacteria and eukaryotes have multiple error-prone DNA polymerases that have the ability to synthesize past a lesion on the template . These enzymes have this ability because they are not constrained to follow standard base pairing rules. Note that this DNA synthesis is not to repair the lesion, but simply to bypass it, to continue replication.
That will allow the cell to return to the lesion to repair it. FIGURE 1. compares an advancing replication fork with what happens when there is damage to a base in the DNA or a nick in one strand. In either case, DNA synthesis is halted, and the replication fork either is stalled or is disrupted and collapses.
Replication-fork stalling appears to be quite common; estimates for the frequency in E. coli suggest that 18%–50% of bacteria encounter a problem during a replication cycle. E. coli has two error-prone DNA polymerases that can replicate through a lesion, DNA polymerases IV and V (see the chapter titled Repair Systems), plus the repair DNA polymerase II, that are used for translesion synthesis. Eukaryotes have five error-prone DNA polymerases with different specificities.
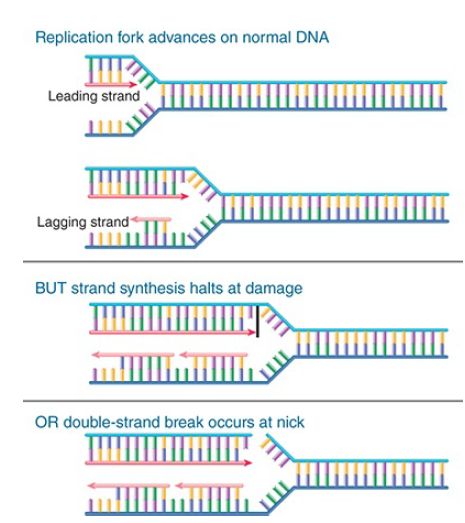
FIGURE 1. The replication fork stalls and may collapse when it reaches a damaged base or a nick in DNA. Arrowheads indicate 3′ ends.
There are two consequences when lesion bypass occurs. First, when the replication complex stalls at a lesion, the polymerase on the strand with the lesion must be removed from the template and replaced by an error-prone polymerase. Second, when the damage has been bypassed, the repair polymerase must be removed and the replication complex reinserted. When used for lesion bypass during replication, these error-prone DNA polymerases replace the replisome and are connected to the PCNA clamp temporarily to allow the lesion bypass polymerase to insert nucleotides opposite the lesion. DNA polymerase III then replaces the error-prone polymerase. The consequences can be different, depending on whether the lesion has occurred on the lagging or leading strand. The replication polymerase on the lagging strand might be more easily replaced.
Alternatively, the situation can be rescued by a recombination event that excises and replaces the damage or provides a new duplex to replace the region containing the double-strand break. The principle of the repair event is to use the built-in redundancy of information between the two DNA strands. FIGURE 2 shows the key events in such a repair event. Basically, information from the undamaged DNA daughter duplex is used to repair the damaged sequence. This creates a typical recombination junction that is resolved by the same systems that perform homologous recombination. In fact, one view is that the major importance of these systems for the cell is in repairing damaged DNA at stalled replication forks.
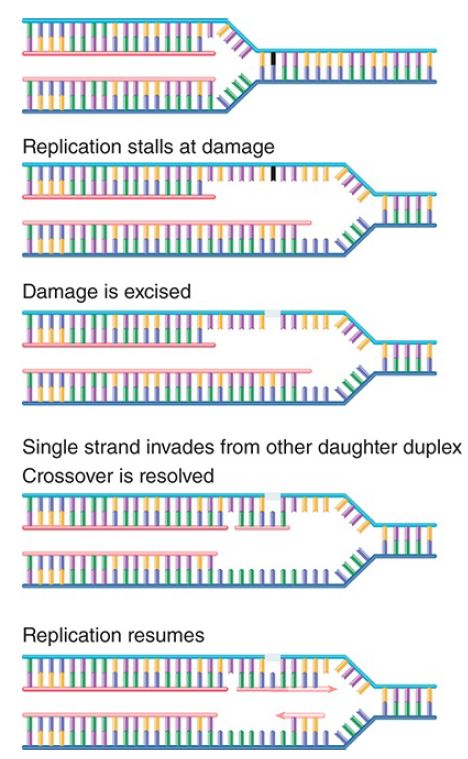
FIGURE 2. When replication halts at damaged DNA, the damaged sequence is excised and the complementary (newly synthesized) strand of the other daughter duplex crosses over to repair the gap. Replication can now resume, and the gaps are filled in.
After the damage has been repaired, the replication fork must be restarted. FIGURE 3. shows that this can be accomplished by assembly of the primosome, which in effect reloads DnaB so that helicase action can continue. Early work on replication made extensive use of phage ΦX174 and led to the discovery of a complex system for priming. A primosome assembles at a unique phage site on its single-stranded DNA called the assembly site (pas). The pas is the equivalent of an origin for synthesis of the complementary strand of ΦX174. The primosome consists of six proteins: PriA, PriB, PriC, DnaT, DnaB, and DnaC. Two alternative assembly pathways exist, one beginning with PriA and the other with PriC. This might reflect the many types of DNA damage that can occur.
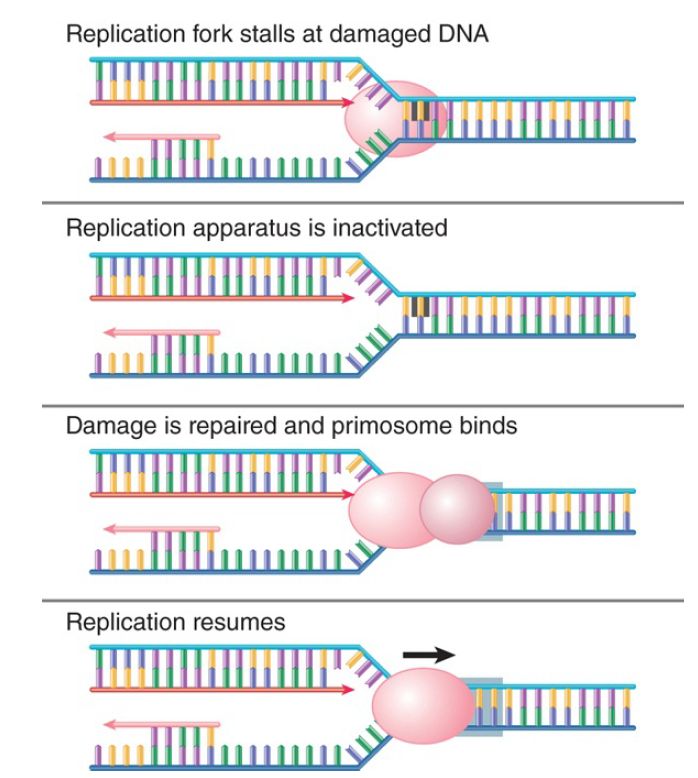
FIGURE 3. The primosome is required to restart a stalled replication fork after the DNA has been repaired.
On ΦX174 DNA, the primosome forms initially at the pas; primers are subsequently initiated at a variety of sites. PriA translocates along the DNA, displacing SSB, to reach additional sites at which priming occurs. As in the E. coli oriC replicon, DnaB plays a key role in unwinding and priming in ΦX174 replicons. The role of PriA is to load DnaB, which in turn recruits DnaG primase to prime DNA synthesis for the conversion of single-stranded viral DNA to the double-stranded DNA form.
It has always been puzzling that when replicating in E. coli, ΦX174 origins should use a complex structure that is not required to replicate the bacterial chromosome. Why does the bacterium provide this complex? The answer is provided by the fate of the stalled replication fork. The mechanism used at oriC is specific for origin DNA sequence and cannot be used to restart replication following lesion bypass because each lesion occurs in a different sequence. A separate mechanism employing structural rather than sequence recognition is used.
The proteins encoded by the E. coli pri genes form the core of theprimo some. ΦX174 has simply co-opted the primosome for its own replication. The PriA DNA helicase binds first to the single-strand region in cooperation with SSB. The key event in localizing the primosome is the ability of PriA to displace SSB from singlestranded DNA. PriA then recruits PriB and DnaT, which is then able to recruit the DnaB/C complex as described earlier . The alternate replisome loading system only requires Pri C. Replication fork reactivation is a common (and therefore important)reaction. It can be required in most chromosomal replication cycles. It is impeded by mutations in either the retrieval systems that replace the damaged DNA or in the components of the primosome.



|
|
|
|
تفوقت في الاختبار على الجميع.. فاكهة "خارقة" في عالم التغذية
|
|
|
|
|
|
|
أمين عام أوبك: النفط الخام والغاز الطبيعي "هبة من الله"
|
|
|
|
|
|
|
المجمع العلمي ينظّم ندوة حوارية حول مفهوم العولمة الرقمية في بابل
|
|
|