


 النبات
النبات
 الحيوان
الحيوان
 الأحياء المجهرية
الأحياء المجهرية
 علم الأمراض
علم الأمراض
 التقانة الإحيائية
التقانة الإحيائية
 التقنية الحيوية المكروبية
التقنية الحيوية المكروبية
 التقنية الحياتية النانوية
التقنية الحياتية النانوية
 علم الأجنة
علم الأجنة
 الأحياء الجزيئي
الأحياء الجزيئي
 علم وظائف الأعضاء
علم وظائف الأعضاء
 الغدد
الغدد
 المضادات الحيوية
المضادات الحيوية|
Read More
Date: 17-5-2021
Date: 28-4-2016
Date: 3-12-2015
|
Lambda Recombination Occurs in an Intasome
KEY CONCEPTS
- Lambda integration takes place in a large complex that also includes the host protein IHF.
- The excision reaction requires Int and Xis and recognizes the ends of the prophage DNA as substrates.
Unlike the Cre/lox recombination system, which requires only the enzyme and the two recombining sites, phage lambda recombination occurs in a large structure and has different components for each direction of the reaction (integration versus excision).
The host protein IHF is required for both integration and excision. IHF is a 20-kD protein of two different subunits, which are encoded by the genes himA and himD. IHF is not an essential protein in E. coli and is not required for homologous bacterial recombination. It is one of several proteins with the ability to wrap DNA on a surface.
Mutations in the him genes prevent lambda site–specific recombination and can be suppressed by mutations in λint, which suggests that IHF and Int interact. Site-specific recombination can
be performed in vitro by Int and IHF.
The in vitro reaction requires supercoiling in attP, but not in attB. When the reaction is performed in vitro between two supercoiled DNA molecules, almost all of the supercoiling is retained by the products. Thus, there cannot be any free intermediates in which strand rotation could occur. This was one of the early hints that the reaction proceeds through a Holliday junction. We now know that the reaction proceeds by the mechanism typical of this class of enzymes, which is related to the topoisomerase I mechanism .
Int has two different modes of binding. The C-terminal domain behaves like the Cre recombinase. It binds to inverted sites at the core sequence, positioning itself to make the cleavage and ligation reactions on each strand at the positions illustrated in FIGURE 1. The N-terminal domain binds to sites in the arms of attP that have a different consensus sequence. This binding is responsible for the aggregation of subunits into the intasome. The two domains probably bind DNA simultaneously, thus bringing the arms of attP close to the core.

FIGURE 1. Int and IHF bind to different sites in attP. The Int recognition sequences in the core region include the sites of cutting.
IHF binds to sequences of about 20 bp in attP. The IHF-binding sites are approximately adjacent to sites where Int binds. Xis binds to two sites located close to one another in attP, so that the protected region extends over 30 to 40 bp. Together, Int, Xis, and IHF cover virtually all of attP. The binding of Xis changes the organization of the DNA so that it becomes inert as a substrate for the integration reaction.
When Int and IHF bind to attP, they generate a complex in which all the binding sites are pulled together on the surface of a protein. Supercoiling of attP is needed for the formation of this intasome. The only binding sites in attB are the two Int sites in the core. Int does not bind directly to attB in the form of free DNA, though. The intasome is the intermediate that “captures” attB, as indicated schematically in FIGURE 2.
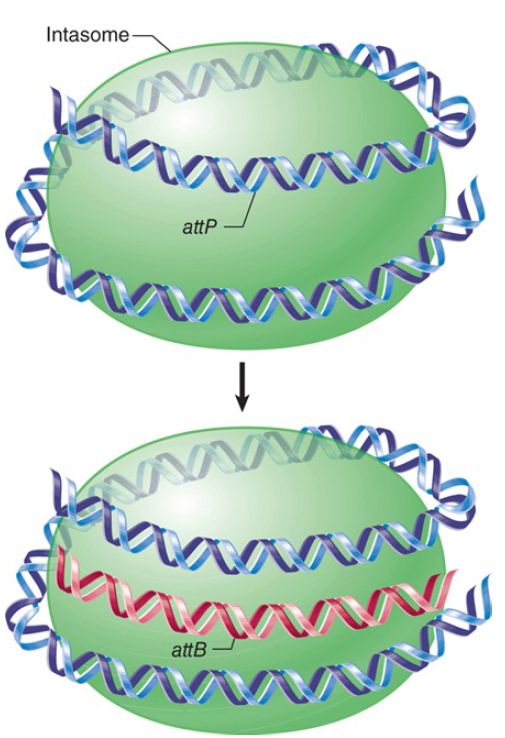
FIGURE 2. Multiple copies of Int protein may organize attP into an intasome, which initiates site-specific recombination by recognizing attB on free DNA.
According to this model, the initial recognition between attP and attB does not depend directly on DNA homology, but instead is determined by the ability of Int proteins to recognize both att
sequences. The two att sites then are brought together in an orientation predetermined by the structure of the intasome. Sequence homology becomes important at this stage, when it is required for the strand-exchange reaction.
The asymmetry of the integration and excision reactions is shown by the fact that Int can form a similar complex with attR only if Xis is added. This complex can pair with a condensed complex that Int forms at attL. IHF is not needed for this reaction. A significant difference between lambda integration/excision and the recombination reactions catalyzed by Cre or Flp is that Intcatalyzed reactions bind the regulatory sequences in the arms of the target sites, bending the DNA and allowing interactions between arm and core sites that drive each reaction to its conclusion. This is why each lambda reaction is irreversible, whereas recombination catalyzed by Cre or Flp is reversible.
Crystal structures of λ-Int tetramers show that, like other recombinases, the tetramer has two active and two inactive subunits that switch roles during recombination. Allosteric interactions triggered by arm-binding control structural transitions in the tetramer that drive the reaction.
Much of the complexity of site-specific recombination may be caused by the need to regulate the reaction so that integration occurs preferentially when the virus is entering the lysogenic state, whereas excision is preferred when the prophage is entering the lytic cycle. By controlling the amounts of Int and Xis, the appropriate reaction will occur.



|
|
|
|
"عادة ليلية" قد تكون المفتاح للوقاية من الخرف
|
|
|
|
|
|
|
ممتص الصدمات: طريقة عمله وأهميته وأبرز علامات تلفه
|
|
|
|
|
|
|
المجمع العلمي للقرآن الكريم يقيم جلسة حوارية لطلبة جامعة الكوفة
|
|
|