


 النبات
النبات
 الحيوان
الحيوان
 الأحياء المجهرية
الأحياء المجهرية
 علم الأمراض
علم الأمراض
 التقانة الإحيائية
التقانة الإحيائية
 التقنية الحيوية المكروبية
التقنية الحيوية المكروبية
 التقنية الحياتية النانوية
التقنية الحياتية النانوية
 علم الأجنة
علم الأجنة
 الأحياء الجزيئي
الأحياء الجزيئي
 علم وظائف الأعضاء
علم وظائف الأعضاء
 الغدد
الغدد
 المضادات الحيوية
المضادات الحيوية|
Read More
Date: 21-10-2015
Date: 22-10-2015
Date: 2-11-2015
|
Protein Synthesis
Proteins are the workhorses of the cell, controlling virtually every reaction within as well as providing structure and serving as signals to other cells. Proteins are long chains of amino acids, and the exact sequence of the amino acids determines the final structure and function of the protein. Instructions for that sequence are encoded in genes. To make a particular protein, a messenger ribonucleic acid (mRNA) copy is made from the gene (in the process called transcription), and the mRNA is transported to the ribosome. Protein synthesis, also called translation, begins when the two ribosomal subunits link onto the mRNA. This step, called initiation, is followed by elongation, in which successive amino acids are added to the growing chain, brought in by transfer RNAs (tRNAs). In this step, the ribosome reads the nucleotides of mRNA three by three, in units called codons, and matches each to three nucleotides on the tRNA, called the anticodon. Finally, during termination, the ribosome unbinds from the mRNA, and the amino acid chain goes on to be processed and folded to make the final, functional protein.
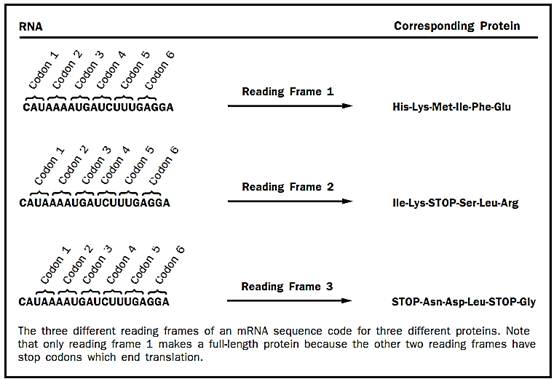
Figure 1
Initiation
In the first step, initiation, the ribosome must bind the mRNA and find the appropriate place to start translating it to make the protein. If the ribosome starts translating the mRNA in the wrong place, the wrong protein will be synthesized. This is a particularly tricky problem because there are three different reading frames in which an mRNA can be read. Each unit of the genetic code, called a codon, is made up of three bases and codes for one amino acid. Completely different protein sequences will be read out by the ribosome if it starts translating with the start of the first codon at base 0, base 1, or base 2 (Figure 1). Thus, it is easy to see why the ribosome must have a way to find the correct starting point for translating each different mRNA.
In almost every known case, translation begins at the three-base codon those codes for the amino acid methionine. This codon has the sequence AUG. Ribosomes are made up of two parts, called subunits, that contain both protein and RNA components. It is the job of the smaller ribosomal subunit to locate the AUG codon that will be used as the starting point for translation (called the initiation codon). Although always starting at AUG helps solve the reading frame problem, finding the right AUG is not an entirely straightforward task. There is often more than one AUG codon in an mRNA, and the small ribosomal subunit must find the correct one if the right protein is to be made.
Initiation in Prokaryotes. In prokaryotes (bacteria) there is a nucleotide sequence on the upstream (5-prime, or 5') side of the initiation codon that tells the ribosome that the next AUG sequence is the correct place to start translating the mRNA. This sequence is called the Shine-Delgarno sequence, after its discoverers. The Shine-Delgarno sequence forms base pairs with RNA in the small ribosomal subunit, thus binding the ribosomal subunit to the mRNA near the initiation codon.
Next, a special tRNA forms base pairs with the AUG sequence of the initiation codon. The tRNA contains the complementary sequence to AUG as its anticodon. This tRNA carries a modified version of the amino acid methionine (fMet-tRNAi or formylmethionyl initiator tRNA) and is already bound to the small ribosomal subunit. The interaction of codon and anticodon triggers a series of events that is not entirely understood but those results in the joining of the large ribosomal subunit to the small ribosomal subunit. The resulting complex is called an initiation complex; it is a whole ribosome bound to an mRNA and an initiator tRNA, positioned so as to make the correct protein from the mRNA.
Initiation in Eukaryotes. In eukaryotes (animals, plants, fungi, and pro- tists), the Shine-Delgarno sequence is missing from the small ribosomal subunit’s RNA, and thus a different mechanism is used for locating the initiation codon. The strategy employed by eukaryotes is more complex and less well understood than that used by prokaryotes. In eukaryotes, the small ribosomal subunit is thought to bind to the 5' end of the mRNA. This binding is mediated by a special structure on the 5' end of eukaryotic mRNAs called a 7-methylguanosine cap and is also aided by a special tail of adenosine bases (the poly-A tail) on the 3' end, both of which are added during RNA processing. A group of proteins called initiation factors binds to the 7-methylguanosine cap and poly(A) tail and appears to direct the binding of the small ribosomal subunit to the mRNA near the cap structure.
Once this has happened, the small ribosomal subunit can read along the mRNA and look for an AUG codon, a process called scanning. Recognition of the initiation codon is largely mediated by base-pairing interactions be-tween the AUG codon and the anticodon sequence in a methionyl initiator tRNA (Met-tRNAi; the methionine is not modified with a formyl group in eukaryotes as it is in prokaryotes). As in prokaryotes, this Met-tRNA is al-ready bound to the small ribosomal subunit.
In most cases, the first AUG codon in a eukaryotic mRNA is used as the initiation codon, thus the small subunit locates the correct initiation codon simply by scanning along the mRNA starting at the 5' end until it reaches the first AUG codon. However, the initiation AUG codon may be flanked by certain base sequences not found around other AUG codons not used for initiation. This preferred set of bases around the initiation codon is called the Kozak sequence, named after its discoverer, Marilyn Kozak. How the Kozak sequence helps direct the small ribosomal subunit to use one AUG codon instead of another is not known. As is the case in prokaryotes, once the correct AUG codon has been found, a complex series of steps takes place that results in the joining of the large ribosomal subunit to the small ribosomal subunit to produce an initiation complex: a complete ribosome assembled at the correct place on an mRNA with an initiator tRNA bound to it.
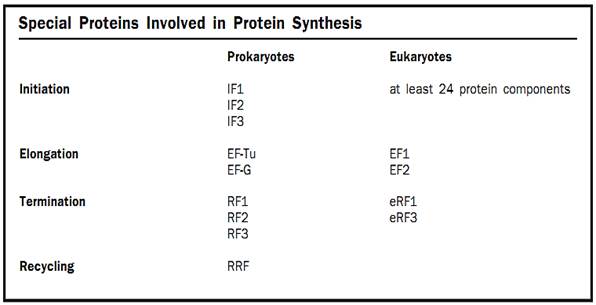
In both prokaryotes and eukaryotes there are proteins called initiation factors that are required for the correct assembly of an initiation complex. In prokaryotes there are three initiation factors, logically enough called IF1, IF2, and IF3. IF2 helps the fMet-tRNAi bind to the small ribosomal subunit. IF3’s main role appears to be to ensure that an AUG, and not another codon, is used as the starting site of translation. That is, IF3 monitors the fidelity of the selection of the initiation codon. IF1 appears to prevent the initiator tRNA from binding to the wrong place in the small ribosomal subunit.
In eukaryotes, the situation is considerably more complex, with at least twenty-four protein components required for the initiation process.
Elongation
In the next phase of protein synthesis, elongation, the ribosome joins amino acids together in the sequence determined by the mRNA to make the corresponding protein. Amino acids are brought onto the ribosome attached to tRNAs. tRNAs are the adapter molecules that allow the ribosome to translate the information contained in the codon sequence of the mRNA into the amino acid sequence of a protein. This decoding happens by base pairing between the anticodon bases of the tRNA and the codon bases of the mRNA. When all three anticodon bases of the tRNA form base pairs with the next codon of the mRNA, the ribosome, with the aid of an elongation factor protein, recognizes that this tRNA has the correct amino acid attached to it and adds this amino acid to the growing protein chain. The process can then be repeated until the entire protein has been synthesized.
As just mentioned, elongation requires the help of elongation factor proteins. The tRNAs with attached amino acids (called aminoacyl tRNAs) are brought onto the ribosome by one such elongation factor. This factor is called EF-Tu in prokaryotes and EF1 in eukaryotes. Its job is to bring aminoacyl tRNAs onto the ribosome and then to help the ribosome make sure that this tRNA has the correct amino acid attached to it. The ribosome has three aminoacyl tRNA binding sites: the acceptor site (A), the peptidyl site (P), and the exit site (E). The tRNA that has the growing protein attached to it binds in the P site (hence the name peptidyl, for peptide). The incoming aminoacyl tRNA, containing the next amino acid to be added, binds in the A site. The A site is where decoding of the genetic code takes place; the correct aminoacyl tRNA is selected to match the next codon of the mRNA. Spent tRNAs that no longer have an amino acid or the growing peptide chain attached to them end up in the E site, from which they fall off the ribosome back into the cytoplasm, where they can pick up new amino acids.
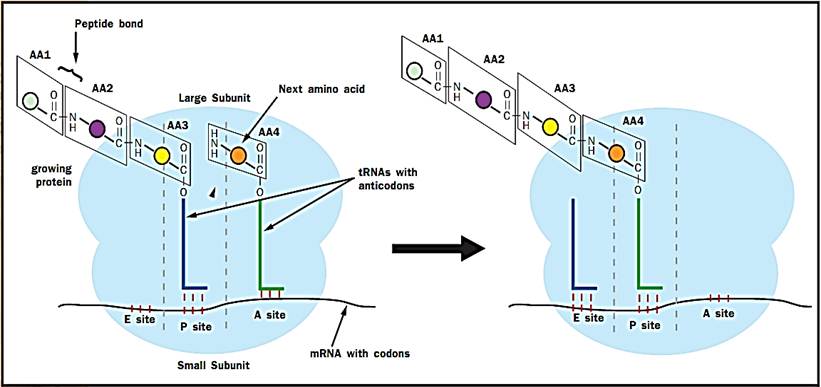
Figure 2. Peptide bond formation by the ribosome. The three lines between the mRNAs and the tRNA indicate base pairing between the codon of the mRNA and the anticodon of the tRNA.
Once the A site is occupied by the correct tRNA, the ribosome links the new amino acid to the growing peptide chain. It does this by catalyzing the formation of a peptide (amide) bond between the amino (NH2) group of the new amino acid in the A site and the carbonyl (CO) group that attaches the growing protein chain to the tRNA in the P site (Figure 2). This results in an intermediate state of the ribosome, called a hybrid state, in which the tRNA in the P site has lost the growing protein chain and moved partially into the E site, and the tRNA in the A site now has the growing protein chain attached to it and has moved partially into the P site.
To complete the round of elongation, a second elongation factor, called EF-G in prokaryotes and EF2 in eukaryotes, is needed. This elongation factor moves the TRNAs such that the spent tRNA that has lost the protein chain moves fully into the E site, and the tRNA with the growing protein chain moves fully into the P site. The mRNA is also shifted over one codon by EF-G, so that the next codon is in the A site. The A site is now empty of tRNAs and the next aminoacyl tRNA can be brought into it.
Many antibiotics (drugs that kill bacteria) affect the elongation phase of prokaryotic translation. Some decrease the fidelity (accuracy) with which the ribosome decodes the mRNA and the wrong amino acids get put into the proteins. This decrease in fidelity leads to an accumulation of proteins that do not work, which eventually kills the bacterium. Other antibiotics prevent the formation of the peptide bond or the movement of the tRNAs by EF-G after the peptide bond has been formed. The reason these drugs are effective on bacteria without killing the patient is that prokaryotic ribosomes have some different structural features than eukaryotic ribosomes, and thus these drugs can bind to the prokaryotic (bacterial) ribosomes but not the eukaryotic (that is, human) ribosomes. Since viruses use human ribosomes to reproduce, these antibiotics are not effective against them.
Termination
The end of the code for the protein in the mRNA is signaled by one of three special codons called stop codons. These stop codons have the sequences UAA, UAG, and UGA. In prokaryotes, the stop codons are bound by one of two release factor proteins (RFs) in prokaryotes: RF1 or RF2. These release factors cause the ribosome to cleave the finished protein off the tRNA in the P site. A third release factor, RF3, is responsible for releasing RF1 and RF2 from the ribosome after they have recognized the stop codon and caused the protein to be cleaved off the tRNA. Eukaryotes appear to have one protein, eRF1, that performs the functions of RF1 and RF2, and a second protein, eRF3, that performs the function of RF3. Once released, the protein can then go on to perform its function in the cell.
After the protein has been cleaved off the tRNA, the two ribosomal sub-units must be dissociated from one another so that the ribosome can start translating another mRNA. This process is called recycling. In prokaryotes, recycling requires three proteins: one initiation factor (IF3), one elongation factor (EF-G), and a ribosome recycling factor called RRF. Once the subunits are dissociated from each other the whole process of translation can begin again.
Protein Folding
A functional protein is not a long, stretched-out chain of amino acids but rather a complex, three-dimensional structure. That is, each protein must fold up into a particular shape, or conformation, in order to perform its function in the cell. The evidence strongly suggests that all of the information required for the protein to fold into its correct three- dimensional structure is contained in the amino acid sequence of the protein (rather than, say , being determined by some other factor in the cell). However, as the pro-tein is being synthesized on the ribosome there is a danger that the unfinished protein will begin to fold up incorrectly because the rest of the protein has not yet been made. It is also possible that the unfinished protein will interact with other unfinished proteins being made on other ribosomes and form what is called an aggregate: a network of partially folded proteins that have interacted with each other rather than with themselves, thus producing a mess inside the cell. Such protein aggregates can be fatal for the cell. It is the job of a class of proteins called chaperones to bind to the growing protein chains as they are synthesized by ribosomes and prevent aggregates from forming or the proteins from folding incorrectly before they have been fully synthesized. Chaperones may also help proteins efficiently fold up into the correct three-dimensional structure once translation is complete.
Protein Modification
While the mRNA encodes the complete amino acid sequence of the corresponding protein, some proteins are altered after they are translated. This process is called post-translational modification. For example, some proteases (proteins that digest other proteins) are synthesized by the ribosome as precursor proteins (pro-proteins) that contain an extra sequence of amino acids at one end that prevents them from digesting any proteins until they get to the right place (usually outside of the cell). Once the proteases reach their destination, the amino acid sequences that prevent them from being active (called pro-sequences) are removed (by another protein), and the proteases can begin digesting other proteins. If these pro-sequences did not ex- ist, the proteases would digest all of the useful proteins inside the cells that made them—which would not be a good thing.
Many proteins made by eukaryotic cells are modified by having sugars attached to various amino acids, a process called glycosylation. Proteins that are destined to be exported from the cell or are going to be inserted into the cell’s membrane enter the endoplasmic reticulum (ER) as they are synthesized by ribosomes that bind to the surface of the ER and feed the new proteins into the ER through small pores. Inside the ER, sugars are added to the protein, which is then sent to the Golgi apparatus where some of the sugars are removed and additional sugars are added. The role of protein glycosylation is not well understood, but because many euykaryotic proteins are glycosylated, it is clearly important.
There are a number of additional ways that proteins can be modified after they are made. For example, many proteins can have one or more phosphate groups added to them by enzymes called kinases. These phosphorylations are often used by the cell to regulate the activity of specific proteins; the phosphorylated form of the protein often has different properties than the unphosphorylated form.
Protein Degradation
When a protein has outlived its usefulness or become damaged, it is degraded by the cell. In eukaryotes, a protein that is to be degraded has a number of copies of the small protein ubiquitin attached to it by a series of ubiquitin-adding enzymes. Ubiquitin serves as a tag that marks the protein for degradation. A tagged protein is then sucked into a large cellular machine called the proteasome, which itself is made up of a number of protein components and looks something like a trash can. Inside the proteasome, the tagged protein is digested into small peptide fragments that are released into the cytoplasm where they can be further digested into free amino acids by other proteases. The life of a protein begins in one cellular machine called the ribosome and ends in another called the proteasome.
References
Lewin, Benjamin. Genes VI. Oxford: Oxford University Press, 1997.
Merrick, William C., and John W. B. Hershey. “The Pathway and Mechanism of Eukaryotic Protein Synthesis.” In Translational Control. Plainview, NY: Cold Spring Harbor Laboratory Press, 1996.
Stryer, Lubert. Biochemistry, 4th ed. New York: W. H. Freeman and Company, 1995.



|
|
|
|
التوتر والسرطان.. علماء يحذرون من "صلة خطيرة"
|
|
|
|
|
|
|
مرآة السيارة: مدى دقة عكسها للصورة الصحيحة
|
|
|
|
|
|
|
نحو شراكة وطنية متكاملة.. الأمين العام للعتبة الحسينية يبحث مع وكيل وزارة الخارجية آفاق التعاون المؤسسي
|
|
|