


 النبات
النبات
 الحيوان
الحيوان
 الأحياء المجهرية
الأحياء المجهرية
 علم الأمراض
علم الأمراض
 التقانة الإحيائية
التقانة الإحيائية
 التقنية الحيوية المكروبية
التقنية الحيوية المكروبية
 التقنية الحياتية النانوية
التقنية الحياتية النانوية
 علم الأجنة
علم الأجنة
 الأحياء الجزيئي
الأحياء الجزيئي
 علم وظائف الأعضاء
علم وظائف الأعضاء
 الغدد
الغدد
 المضادات الحيوية
المضادات الحيوية|
Read More
Date: 1-5-2016
Date: 30-3-2021
Date: 21-3-2021
|
Initiation Is Followed by Promoter Clearance and Elongation
KEY CONCEPTS
- TFII B, TFII E, and TFII H are required to melt DNA to allow polymerase movement.
- Phosphorylation of the carboxy-terminal domain (CTD) is required for promoter clearance and elongation to begin.
- Further phosphorylation of the CTD is required at some promoters to end pausing and abortive initiation.
- The histone octamers must be temporarily modified during the transit of the RNA polymerase.
- The CTD coordinates processing of RNA with transcription.
- Transcribed genes are preferentially repaired when DNA damage occurs.
TFII H provides the link to a complex of repair enzymes.
Promoter melting (DNA unwinding) is necessary to begin the process of transcription. TFIIH is required for the formation of the open complex in conjunction with ATP hydrolysis to provide torsional stress for unwinding. Some final steps are then needed to release the RNA polymerase from the promoter once the first nucleotide bonds have been formed. Promoter clearance is the key regulated step in eukaryotes in determining if a poised gene or an active gene will be transcribed. This step is controlled by enhancers. (Note that the key step in bacterial transcription is conversion of the closed complex to the open complex; see the chapter titled Prokaryotic Transcription.) Most of the general transcription factors are required solely to bind RNA polymerase to the promoter, but some act at a later stage.
The transcription factors that bind enhancers usually do not directly contact elements at the promoter to control it, but rather bind to a coactivator that binds to the promoter elements. The coactivator Mediator is one of the most common coactivators. This is a very large multisubunit protein complex. In multicellular eukaryotes, it can contain 30 subunits or more. Many cell-type and gene-specific forms of Mediator contain a common core of subunits conserved from yeast to humans that integrate signals from many enhancerbound transcription factors. Both poised and active genes require the interaction of the transcription factors bound to enhancers with the promoter by DNA looping with Mediator as the intermediate. The last factors to join the initiation complex are TFII E and TFII H.
They act at the later stages of initiation for unwinding the DNA. Binding of TFII E causes the boundary of the region protected downstream to be extended by another turn of the double helix, to+30. TFII H is the only general transcription factor that has multiple independent enzymatic activities. Its several activities include an ATPase, helicases of both polarities, and a kinase activity that can phosphorylate the CTD tail of RNA polymerase II (on serine 5 of the heptapeptide repeat). TFII H is an exceptional factor that may also play a role in elongation. Its interaction with DNA downstream of the start point is required for RNA polymerase to escape from the promoter. TFII H is also involved in repair of damage to DNA .
On a linear template, ATP hydrolysis, TFII E, and the helicase activity of TFII H (provided by the XPB and XPD subunits) are required for polymerase movement. This requirement is bypassed with a supercoiled template. This suggests that TFII E and TFII H are required to melt DNA to allow polymerase movement to begin. The helicase activity of the XPB subunit of TFII H is responsible for the actual melting of DNA.
RNA polymerase II stutters when it starts transcription. (The result is not dissimilar to the abortive initiation of bacterial RNA polymerase discussed in the chapter titled Prokaryotic Transcription, although the mechanism is different.) RNA polymerase II terminates after a short distance; small oligonucleotides of 4 to 5 nucleotides are unstable; and the crystal structures of these RNA–DNA hybrids are unordered. Only longer hybrids have proper base pairing. The short RNA products are degraded rapidly. The suggestion is that this abortive initiation is a form of promoter proofreading. To extend elongation into the transcription unit, a kinase complex, P-TEFb, is required. P-TEFb contains the CDK9 kinase, which is a member of the kinase family that controls the cell cycle. P-TEFb acts on the CTD to phosphorylate it further (on serine 2 of the heptapeptide repeat). It is not yet understood why this effect is required at some promoters but not others or how it is regulated.
Phosphorylation of the CTD tail is needed to release RNA polymerase II from the promoter and transcription factors so that it can make the transition to the elongating form, as shown in FIGURE 1. Real-time observation of live cells shows a bursting pattern that is gene specific, rather than continuous initiation. The phosphorylation pattern on the CTD is dynamic during the elongation process, catalyzed and controlled by multiple protein kinases, including P-TEFb, and phosphatases. Most of the basal transcription factors are released from the promoter at this stage.
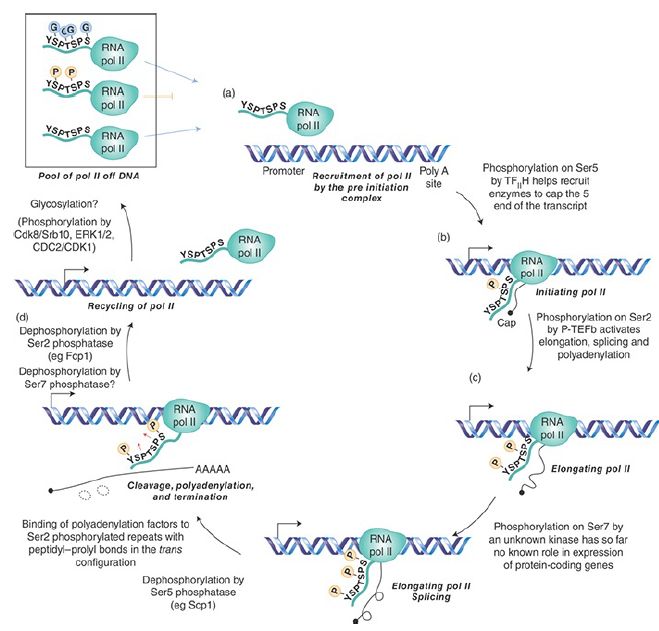
FIGURE 1. Modification of the RNA polymerase II CTD heptapeptide during transcription. The CTD of RNA polymerase II when it enters the preinitiation complex is unphosphorylated. Phosphorylation of Ser residues serves as binding sites for both mRNA processing enzymes and kinases that catalyze further phosphorylation as described in the figure. Reprinted from Trends Genet., vol. 24, S. Egloff and S. Murphy, Cracking the RNA polymerase II CTD code, pp. 280–288. Copyright 2008, with permission from Elsevier [http://www.sciencedirect.com/science/journal/01689525].
The CTD is involved, directly or indirectly, in processing mRNA while it is being synthesized and after it has been released by RNA polymerase II. Sites of phosphorylation on the CTD serve as a recognition or anchor point for other proteins to dock with the polymerase. The capping enzyme (guanylyl transferase), which adds the G residue to the 5′ end of newly synthesized mRNA, binds to CTD phosphorylated at serine 5, the first phosphorylation event catalyzed by TF H. This may be important in enabling it to modify (and thus protect) the 5′ end as soon as it is synthesized.
Subsequently, serine 2 phosphorylation by P-TEFb leads to recruitment of a set of proteins called SCAFs to the CTD, and they, in turn, bind to splicing factors. This may be a means of coordinating transcription and splicing. Some components of the cleavage/polyadenylation apparatus used during transcription termination also bind to the CTD phosphorylated at serine 2. Oddly enough, they do so at the time of initiation, so that RNA polymerase is ready for the 3′ end processing reactions as soon as it sets out.
Finally, export from the nucleus through the nuclear pore is also controlled by the CTD and may be coordinated with 3′ end processing. All of this suggests that the CTD may be a general focus for connecting other processes with transcription. In the cases of capping and splicing, the CTD functions indirectly to promote formation of the protein complexes that undertake the reactions. In the case of 3′ end generation, it may participate directly in the reaction. Control of the life history of an mRNA does not end here. Recent data show that in yeast a subset of mRNAs exist whose cytoplasmic stability or turnover is directly controlled by the promoter/upstream activating sequence (UAS). Binding sites for specific transcription factors control recruitment of stability/instability factors that bind to the mRNA during transcription.
The key event in determining whether (and when, in the case of a poised or paused polymerase, see the following discussion) a gene will be expressed is promoter clearance, release from the promoter regulated by PAF-1, the gatekeeper for regulation of gene expression. Once that has occurred and initiation factors are released, there is a transition to the elongation phase. The transcription complex now consists of the RNA polymerase II, the basal factors TFII E and TFII H, and all of the enzymes and factors bound to the CTD. Elongation factors such as TFII F and TFII S and others to prevent inappropriate pausing may be present in another large complex called super elongation complex (SEC).
The RNA polymerase, like the ribosome, functions as a Brownian ratchet where random fluctuations are stabilized and (usually) converted into forward motion by the binding of nucleotides. This,then, mean s that forward as well as backward or backtracking motion occurs. Backtracking also occurs when an incorrect nucleotide is inserted and the duplex structure of the 3′ end is improperly base paired. Backtracking is a necessary component of the fidelity mechanism. The dynamics of this are controlled by the underlying DNA sequence context and elongation factors such as TFII F, TFII S, Elongin, and a number of others.
As discussed earlier in the section The Basal Apparatus Assembles at the Promoter, considerable heterogeneity can exist in the DNA sequence elements that comprise the core promoter that can lead to promoter specificity of different genes. One of these elements is known as the pause button, a G-C–rich sequence typically located downstream from the start of initiation. This element has been found in a surprising number of Drosophila developmental genes, among others. Release from pausing requires a separate set of regulatory steps controlled by the gene’s enhancer and a 7SK snRNA that provides a link between the enhancer, the polymerase, and a required chromatin mark. P-TEFb is required to phosphorylate negative regulating pause factors in order to inactivate them and to phosphorylate the CTD for release.
A subset of human genes in a paused state is regulated by the oncogene transcription factor cMyc . P-TEFb is specifically recruited to these genes by cMyc in order to release them from the paused state.
In summary, the general process of initiation is similar to that catalyzed by bacterial RNA polymerase. Binding of RNA polymerase generates a closed complex, which is converted at a
later stage to an open complex in which the DNA strands have been separated. In the bacterial reaction, formation of the open complex completes the necessary structural change to DNA; a difference in the eukaryotic reaction is that further unwinding of the template is needed after this stage.
This complex now has to transcribe a chromatin template, through nucleosomes. The whole gene may be in open chromatin, especially if it is not too large, or only the area around the promoter. Some genes, like the Duchenne muscular dystrophy gene (DMD), can be megabases in size and require many hours to transcribe. The histone octamers must be transiently modified—in some cases temporarily disassembled—and then reassembled on the template (see the chapters titled Chromatin and Eukaryotic Transcription Regulation for more details). The octamer itself is changed by this process, having some of the canonical histone H3 replaced by the variant H3.3 during active transcription. A model exists in which the first polymerase to leave the promoter acts as a pathfinder polymerase. Its major function is to ensure that the entire gene is in open chromatin. It carries with it enzyme complexes to facilitate transcription through nucleosomes. Both the initiation factor TFII F and the elongation factor TFII S are required. Histone H2B is dynamically monoubiquitinated in actively transcribed chromatin. This is required in order for the second step, methylation of histone H3, which is, in turn, required for the recruitment of chromatin remodelers .
The most recent model has each polymerase using a chromatinremodeling complex together with a histone chaperone to remove an H2A–H2B dimer, leaving a hexamer (in place of the octamer), which is easier to temporarily displace. These modifications also are necessary to reassemble the nucleosome octamer on the DNA in the wake of the RNA polymerase . In both bacteria and eukaryotes, there is a direct link from RNA polymerase to the activation of DNA repair. The basic phenomenon was first observed because transcribed genes are preferentially repaired. It was then discovered that it is only the template strand of DNA that is the target—the nontemplate strand is repaired at the same rate as bulk DNA. When RNA polymerase encounters DNA damage in the template strand, it stalls because it cannot use the damaged sequences as a template to direct complementary base pairing. This explains the specificity of the effect for the template strand (damage in the nontemplate strand does not impede progress of the RNA polymerase). Stalled polymerase at a damage site recruits a pair of proteins, CSA and CSB (proteins with the name CS are encoded by genes in which mutations lead to the disease Cockayne syndrome). The general transcription factor TFII H, already present with the elongating polymerase, is essential to the repair process. TFII H is found in alternative forms, which consist of a core associated with other subunits.
TFII H has a common function in both initiating transcription and repairing damage. The same TFII H helicase subunits (XPB and XPD) create the initial transcription bubble and melt DNA at a damaged site. Subunits with the name XP are encoded by genes in which mutations cause the disease xeroderma pigmentosum, which causes a predisposition to cancer. The role of TFII H subunits in DNA repair is discussed in detail in the Repair Systems chapter.
The repair function may require modification or degradation of a stalled RNA polymerase. The large subunit of RNA polymerase is degraded by the ubiquitylation pathway when the enzyme stalls at sites of ultraviolet (UV) damage. The connection between the transcription/repair apparatus as such and the degradation of RNA polymerase is not yet fully understood. It is possible that removal of the polymerase is necessary once it has become stalled.



|
|
|
|
تفوقت في الاختبار على الجميع.. فاكهة "خارقة" في عالم التغذية
|
|
|
|
|
|
|
أمين عام أوبك: النفط الخام والغاز الطبيعي "هبة من الله"
|
|
|
|
|
|
|
المجمع العلمي ينظّم ندوة حوارية حول مفهوم العولمة الرقمية في بابل
|
|
|