


 النبات
النبات
 الحيوان
الحيوان
 الأحياء المجهرية
الأحياء المجهرية
 علم الأمراض
علم الأمراض
 التقانة الإحيائية
التقانة الإحيائية
 التقنية الحيوية المكروبية
التقنية الحيوية المكروبية
 التقنية الحياتية النانوية
التقنية الحياتية النانوية
 علم الأجنة
علم الأجنة
 الأحياء الجزيئي
الأحياء الجزيئي
 علم وظائف الأعضاء
علم وظائف الأعضاء
 الغدد
الغدد
 المضادات الحيوية
المضادات الحيوية|
Read More
Date: 1-3-2021
Date: 16-5-2021
Date: 4-5-2016
|
Satellite DNAs Often Lie in Heterochromatin
KEY CONCEPTS
-Highly repetitive DNA (or satellite DNA) has a very short repeating sequence and no coding function.
-Satellite DNA occurs in large blocks that can have distinct physical properties.
-Satellite DNA is often the major constituent of centromeric heterochromatin.
Repetitive DNA is characterized by its relatively rapid rate of renaturation. The component that renatures most rapidly in a eukaryotic genome is called highly repetitive DNA and consists of very short sequences repeated many times in tandem in large clusters. As a result of its short repeating unit, it is sometimes described as simple sequence DNA. This component is present in almost all multicellular eukaryotic genomes, but its overall amount is extremely variable. In mammalian genomes it is typically less than 10%, but in (for example) the fruit fly Drosophila virilis, it amounts to about 50%. In addition to the large clusters in which this type of sequence was originally discovered, there are smaller clusters interspersed with nonrepetitive DNA. It typically consists of short sequences that are repeated in identical or related copies in the genome.
In addition to simple sequence DNA, multicellular eukaryotes have complex satellites with longer repeat units, usually in heterochromatin (but sometimes in euchromatic) regions. For example, Drosophila species have the 1.688 g-cmr-3 class of satellite DNA that consists of a 359-bp repeat unit. In humans, the α satellite family, found in centromeric regions, has a repeat unit length of 171 bp. The human β satellite family has 68-bp repeat units interspersed with a longer 3.3-kb repeat unit that includes pseudogenes.
The tandem repetition of a short sequence often has distinctive physical properties that researchers can use to isolate it. In some cases, the repetitive sequence has a base composition distinct from the genome average, which allows it to form a separate fraction by virtue of its distinct buoyant density. A fraction of this sort is called satellite DNA. The term satellite DNA is essentially synonymous with simple sequence DNA. Consistent with its simple sequence, this DNA might or might not be transcribed, but it is not translated. (In some species, there is evidence that short RNAs are required for heterochromatin formation, suggesting that there is
transcription of sequences in heterochromatic regions of chromosomes, which contain satellite DNA.
Tandemly repeated sequences are especially liable to undergo misalignments during chromosome pairing, and therefore the sizes of tandem clusters tend to be highly polymorphic, with wide variations between individuals. In fact, the smaller clusters of such sequences can be used to characterize individual genomes in the technique of “DNA profiling” .
The buoyant density of a duplex DNA depends on its GC content according to the empirical formula:
ρ = 1.660 + 0.00098 (%GC) g-cm−3
Buoyant density is usually determined by centrifuging DNA through a density gradient of cesium chloride (CsCl). The DNA forms a band at the position corresponding to its own density. Fractions of DNA differing in GC content by more than 5% can usually be separated on a density gradient.
When eukaryotic DNA is centrifuged on a density gradient, two categories of DNA may be distinguished:
- Most of the genome forms a continuum of fragments that appear as a rather broad peak centered on the buoyant density corresponding to the average GC content of the genome. This is called the main band.
- Sometimes an additional, smaller peak (or peaks) is seen at a different value. This material is the satellite DNA.
Satellites are present in many eukaryotic genomes. They can be either heavier or lighter than the main band, but it is uncommon for them to represent more than 5% of the total DNA. A clear example is provided by mouse DNA, as shown in FIGURE 1. The graph is a quantitative scan of the bands formed when mouse DNA is centrifuged through a CsCl density gradient. The main band contains 92% of the genome and is centered on a buoyant density of 1.701 g-cm−3 (corresponding to its average GC content of 42%, typical for a mammal). The smaller peak represents 8% of the genome and has a distinct buoyant density of 1.690 g-cm−3 . It contains the mouse satellite DNA, whose GC content (30%) is much lower than any other part of the genome.
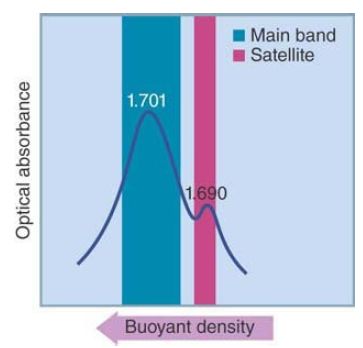
FIGURE 1 Mouse DNA is separated into a main band and a satellite band by centrifugation through a density gradient of CsCl.
The behavior of satellite DNA in density gradients is oftenanomalous. When the actual base composition of a satellite is determined, it is different from the prediction based on its buoyant density. The reason is that ρ is a function not just of base composition but also of the constitution in terms of nearest neighbor pairs. For simple sequences, these are likely to deviate
from the random pairwise relationships needed to obey the equation for buoyant density. In addition, satellite DNA can be methylated, which changes its density.
Often, most of the highly repetitive DNA of a genome can be isolated in the form of satellites. When a highly repetitive DNA component does not separate as a satellite, on isolation its properties often prove to be similar to those of satellite DNA. That is to say, highly repetitive DNA consists of multiple tandem repeats with anomalous centrifugation. Material isolated in this manner is sometimes referred to as a cryptic satellite. Together the cryptic and apparent satellites usually account for all the large tandemly repeated blocks of highly repetitive DNA. When a genome has more than one type of highly repetitive DNA, each exists in its own satellite block (although sometimes different blocks are adjacent).
Where in the genome are the blocks of highly repetitive DNA located? An extension of nucleic acid hybridization techniques allows the location of satellite sequences to be directly determined in the chromosome complement. In the technique of in situ hybridization, the chromosomal DNA is denatured by treating cells that have been “squashed.” Next, a solution containing a labeled single-stranded DNA or RNA probe is added. The probe hybridizes with its complementary sequences in the denatured genome.
Researchers can determine the location of the sites of hybridization by a technique to detect the label, such as autoradiography or fluorescence.
Satellite DNAs are found in regions of heterochromatin. Heterochromatin is the term used to describe regions of chromosomes that are permanently tightly coiled up and inert, in contrast with the euchromatin that represents the active component of the genome (see the Chromosomes chapter). Heterochromatin is commonly found at centromeres (the regions where the kinetochores are formed at mitosis and meiosis for controlling chromosome segregation). The centromeric location of satellite DNA suggests that it has some structural function in the chromosome. This function could be connected with the process of chromosome segregation.
FIGURE 2 shows an example of the localization of satellite DNA for the mouse chromosomal complement. In this case, one end of each chromosome is labeled because this is where the centromeres are located in Mus musculus (mouse) chromosomes.
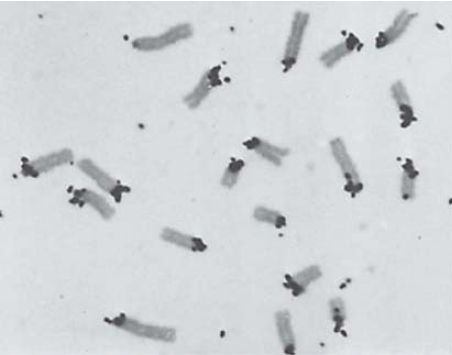
FIGURE 2 Cytological hybridization shows that mouse satellite DNA is located at the centromeres.
Photo courtesy of Mary Lou Pardue and Joseph G. Gall, Carnegie Institution.



|
|
|
|
إجراء أول اختبار لدواء "ثوري" يتصدى لعدة أنواع من السرطان
|
|
|
|
|
|
|
دراسة تكشف "سببا غريبا" يعيق نمو الطيور
|
|
|
|
|
|
قسم الشؤون الفكرية يقيم برنامج (صنّاع المحتوى الهادف) لوفدٍ من محافظة ذي قار
|
|
|
|
الهيأة العليا لإحياء التراث تنظّم ورشة عن تحقيق المخطوطات الناقصة
|
|
|
|
قسم شؤون المعارف يقيم ندوة علمية حول دور الجنوب في حركة الجهاد ضد الإنكليز
|
|
|
|
وفد جامعة الكفيل يزور دار المسنين في النجف الأشرف
|