
 النبات
النبات
 الحيوان
الحيوان
 الأحياء المجهرية
الأحياء المجهرية
 علم الأمراض
علم الأمراض
 التقانة الإحيائية
التقانة الإحيائية
 التقنية الحيوية المكروبية
التقنية الحيوية المكروبية
 التقنية الحياتية النانوية
التقنية الحياتية النانوية
 علم الأجنة
علم الأجنة
 الأحياء الجزيئي
الأحياء الجزيئي
 علم وظائف الأعضاء
علم وظائف الأعضاء
 الغدد
الغدد
 المضادات الحيوية
المضادات الحيوية|
Read More
Date: 14-4-2021
Date:
Date: 19-12-2015
|
Some Exons Correspond to Protein Functional Domains
KEY CONCEPTS
- Proteins can consist of independent functional modules, the boundaries of which, in some cases, correspond to those of exons.
- The exons of some genes appear homologous to the exons of others, suggesting a common exon ancestry.
The issue of the evolution of interrupted genes is more fully considered in the Genome Sequences and Evolution chapter. If proteins evolve by recombining parts of ancestral proteins that were originally separate, the accumulation of protein domains is likely to have occurred sequentially, with one exon added at a time.
Each addition would need to improve upon the advantages of prior additions in a sequence of positive selection events. Are the different function-encoding segments from which these genes might have originally been pieced together reflected in their present structures? If a protein sequence were randomly interrupted, sometimes the interruption would intersect a domain and
sometimes it would lie between domains. If we can associate the functional domains of current proteins with the individual exons of the corresponding genes, this would suggest selective interdomain interruptions rather than random ones.
In some cases, there is a clear relationship between the structures of a gene and its protein product, but these might be special cases. The example par excellence is provided by the immunoglobulin (antibody) proteins—an extracellular system for self-/nonselfdiscrimination that aids in the elimination of foreign pathogens.
Immunoglobulins are encoded by genes in which every exon corresponds exactly to a known functional protein domain. Banks of alternate sequence domains are tapped so that each cell acquires the ability to secrete a cell-specific immunoglobulin with distinctive binding capacity for a foreign antigen that the organism might someday encounter again . FIGURE 1 compares the structure of an immunoglobulin with its gene.

FIGURE 1. Immunoglobulin light chains and heavy chains are encoded by genes whose structures (in their expressed forms) correspond to the distinct domains in the protein. Each protein domain corresponds to an exon; introns are numbered I1 to I5.
An immunoglobulin is a tetramer of two light chains and two heavy chains that covalently bond to generate a protein with several distinct domains. Light chains and heavy chains differ in structure, and there are several types of heavy chains. Each type of chain is produced from a gene that has a series of exons corresponding to the structural domains of the protein.
In many instances, some of the exons of a gene can be identified with particular functions. In secretory proteins, such as insulin, the first exon that encodes the N-terminal region of the polypeptide often specifies a signal sequence needed for transfer across a membrane.
The view that exons are the functional building blocks of genes is supported by cases in which two genes can share some related exons but also have unique exons. FIGURE 2. summarizes the relationship between the receptor for human plasma low-density lipoprotein (LDL) and other proteins. The LDL receptor gene has a series of exons related to the exons of the epidermal growth factor (EGF) precursor gene and another series of exons related to those of the blood protein complement factor C9. Apparently, the LDL receptor gene evolved by the assembly of modules for its various functions. These modules are also used in different combinations in other proteins.
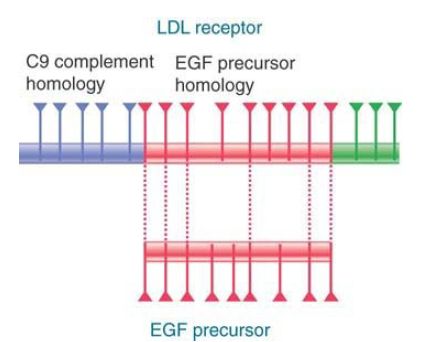
FIGURE 2. The LDL receptor gene consists of 18 exons, some of which are related to EGF precursor exons and some of which are related to the C9 blood complement gene. Triangles mark the positions of introns.
Exons tend to be fairly small—around the size of the smallest polypeptide that can assume a stable folded structure (approximately 20 to 40 residues). It might be that proteins were originally assembled from rather small modules. Each individual module need not correspond to a current function; several modules could have combined to generate a new functional unit. Larger genes tend to have more exons, which is consistent with the view that proteins acquire multiple functions by successively adding appropriate modules.
This suggestion might explain another aspect of protein structure: it appears that the sites represented at exon-intron boundaries often are located at the surface of a protein. As modules are added to a protein, the connections—at least of the most recently added modules—could tend to lie at the surface.



|
|
|
|
التوتر والسرطان.. علماء يحذرون من "صلة خطيرة"
|
|
|
|
|
|
|
مرآة السيارة: مدى دقة عكسها للصورة الصحيحة
|
|
|
|
|
|
|
نحو شراكة وطنية متكاملة.. الأمين العام للعتبة الحسينية يبحث مع وكيل وزارة الخارجية آفاق التعاون المؤسسي
|
|
|