

النبات

مواضيع عامة في علم النبات

الجذور - السيقان - الأوراق

النباتات الوعائية واللاوعائية

البذور (مغطاة البذور - عاريات البذور)

الطحالب

النباتات الطبية


الحيوان

مواضيع عامة في علم الحيوان

علم التشريح

التنوع الإحيائي

البايلوجيا الخلوية


الأحياء المجهرية

البكتيريا

الفطريات

الطفيليات

الفايروسات


علم الأمراض

الاورام

الامراض الوراثية

الامراض المناعية

الامراض المدارية

اضطرابات الدورة الدموية

مواضيع عامة في علم الامراض

الحشرات


التقانة الإحيائية

مواضيع عامة في التقانة الإحيائية


التقنية الحيوية المكروبية

التقنية الحيوية والميكروبات

الفعاليات الحيوية

وراثة الاحياء المجهرية

تصنيف الاحياء المجهرية

الاحياء المجهرية في الطبيعة

أيض الاجهاد

التقنية الحيوية والبيئة

التقنية الحيوية والطب

التقنية الحيوية والزراعة

التقنية الحيوية والصناعة

التقنية الحيوية والطاقة

البحار والطحالب الصغيرة

عزل البروتين

هندسة الجينات


التقنية الحياتية النانوية

مفاهيم التقنية الحيوية النانوية

التراكيب النانوية والمجاهر المستخدمة في رؤيتها

تصنيع وتخليق المواد النانوية

تطبيقات التقنية النانوية والحيوية النانوية

الرقائق والمتحسسات الحيوية

المصفوفات المجهرية وحاسوب الدنا

اللقاحات

البيئة والتلوث


علم الأجنة

اعضاء التكاثر وتشكل الاعراس

الاخصاب

التشطر

العصيبة وتشكل الجسيدات

تشكل اللواحق الجنينية

تكون المعيدة وظهور الطبقات الجنينية

مقدمة لعلم الاجنة


الأحياء الجزيئي

مواضيع عامة في الاحياء الجزيئي


علم وظائف الأعضاء


الغدد

مواضيع عامة في الغدد

الغدد الصم و هرموناتها

الجسم تحت السريري

الغدة النخامية

الغدة الكظرية

الغدة التناسلية

الغدة الدرقية والجار الدرقية

الغدة البنكرياسية

الغدة الصنوبرية

مواضيع عامة في علم وظائف الاعضاء

الخلية الحيوانية

الجهاز العصبي

أعضاء الحس

الجهاز العضلي

السوائل الجسمية

الجهاز الدوري والليمف

الجهاز التنفسي

الجهاز الهضمي

الجهاز البولي


المضادات الميكروبية

مواضيع عامة في المضادات الميكروبية

مضادات البكتيريا

مضادات الفطريات

مضادات الطفيليات

مضادات الفايروسات

علم الخلية

الوراثة

الأحياء العامة

المناعة

التحليلات المرضية

الكيمياء الحيوية

مواضيع متنوعة أخرى

الانزيمات
Histone Acetylation Is Associated with Transcription Activation
المؤلف:
JOCELYN E. KREBS, ELLIOTT S. GOLDSTEIN and STEPHEN T. KILPATRICK
المصدر:
LEWIN’S GENES XII
الجزء والصفحة:
10-6-2021
2708
Histone Acetylation Is Associated with Transcription Activation
KEY CONCEPTS
- Newly synthesized histones are acetylated at specific sites, then deacetylated after incorporation into nucleosomes.
- Histone acetylation is associated with activation of gene expression.
-Transcription activators are associated with histone acetylase activities in large complexes.
- Histone acetyltransferases vary in their target specificity.
- Deacetylation is associated with repression of gene activity.
- Deacetylases are present in complexes with repressor activity.
All of the core histones are subject to multiple covalent modifications, as discussed in the Chromatin chapter. Different modifications result in different functional outcomes. One of the most extensively studied modifications (and the first to be characterized in detail) is lysine acetylation. All core histones are dynamically acetylated on lysine residues in the tails (and occasionally within the globular core). certain patterns of acetylation are associated with newly synthesized histones that are deposited during DNA synthesis in S phase. This specific acetylation pattern is then erased after histones are incorporated into nucleosomes.
Outside of S phase, acetylation of histones in chromatin is generally correlated with the state of gene expression. The correlation was first noticed because histone acetylation is increased in a domain containing active genes, and acetylated chromatin is more sensitive to DNase I. This occurs largely because of acetylation of the nucleosomes (on specific lysines) in the vicinity of the promoter when a gene is activated. The range of nucleosomes targeted for modification can vary. Modification can be a local event—for example, restricted to nucleosomes at a promoter. It can also be a general event, extending over large domains or even to an entire chromosome. Global changes in acetylation occur on sex chromosomes. This is part of the mechanism by which the activities of genes on sex chromosomes are altered to compensate for the presence of two X chromosomes in one sex but only one X chromosome in the other sex . The inactive X chromosome in female mammals has underacetylated histones. The superactive X chromosome in Drosophila males has increased acetylation of H4. This suggests that the presence of acetyl groups may be a prerequisite for a less condensed, active structure. In male Drosophila, the X chromosome is acetylated specifically at K16 of histone H4. The enzyme responsible for this acetylation is called MOF; MOF is recruited to the chromosome as part of a large protein complex. This “dosage compensation” complex is responsible for introducing general changes in the X chromosome that enable it to be more highly expressed. The increased acetylation is only one of its activities.
Acetylation is reversible. Each direction of the reaction is catalyzed by a specific type of enzyme. Enzymes that can acetylate lysine residues in proteins are called histone acetyltransferases (HATs); when these enzymes target lysines in nonhistones, they are also known more generically as lysine (K) acetyltransferases (KATs). The acetyl groups are removed by histone deacetylases (HDACs). HAT enzymes are categorized into two groups: Those in group A act on histones in chromatin and are involved with the control of transcription; those in group B act on newly synthesized histones in the cytosol and are involved with nucleosome assembly.
Two inhibitors have been useful in analyzing acetylation. Trichostatin and butyric acid inhibit histone deacetylases and cause acetylated nucleosomes to accumulate. The use of these inhibitors has supported the general view that acetylation is associated with gene expression; in fact, the ability of butyric acid to cause changes in chromatin resembling those found upon gene activation was one of the first indications of the connection between acetylation and gene activity.
The breakthrough in analyzing the role of histone acetylation was provided by the characterization of the acetylating and deacetylating enzymes and their association with other proteins that are involved in specific events of activation and repression. A basic change in the view of histone acetylation was caused by the discovery that previously identified activators of transcription turned out to also have HAT activity.
The connection was established when the catalytic subunit of a group A HAT was identified as a homolog of the yeast regulator protein Gcn5. It then was shown that yeast Gcn5 itself has HAT activity, with histones H3 and H2B as its preferred substrates in vivo. Gcn5 had previously been identified as part of an adaptor complex required for the function of certain enhancers and their target promoters. It is now known that Gcn5’s HAT activity is required for activation of a number of target genes. Gcn5 was the prototypic HAT that opened the way to the identification of a large family of related acetyltransferase complexes conserved from yeast to mammals. In yeast, Gcn5 is the catalytic subunit of several HAT complexes, including the 1.8-MDa Spt-Ada-Gcn5-acetyltransferase (SAGA) complex, which contains several proteins that are involved in transcription. Among these proteins are several TAF s. In addition, the Taf1 subunit of TFII D is itself an acetyltransferase. Some functional overlap exists between TFII D and SAGA, most notably that yeast can survive the loss of either Taf1 or Gcn5 but cannot tolerate the deletion of both.
This might suggest that an acetyltransferase activity is essential for gene expression, and that it can be provided by either TFII D or SAGA. As might be expected from the size of the SAGA complex, acetylation is only one of its functions. The SAGA complex has histone H2B deubiquitylation activity (dynamic H2B ubiquitylation/deubiquitylation is also associated with transcription), and also contains subunits possessing bromodomains and chromodomains, allowing this complex to interact with acetylated and methylated histones.
One of the first general activators to be characterized as HAT was p300/CREB-binding protein (CBP). (Actually, p300 and CBP are different proteins, but they are so closely related that they are often referred to as a single type of activity.) p300/CBP is a coactivator that links an activator to the basal apparatus . p300/CBP interacts with various activators, including the hormone receptors AP-1 (c-Jun and c-Fos) and MyoD. p300/CBP acetylates multiple histone targets, with a preference for the H4 tail. p300/CBP interacts with another coactivator, PCAF, which is related to Gcn5 and preferentially acetylates H3 in nucleosomes. p300/CBP and PCAF form a complex that functions in transcriptional activation. In some cases yet another HAT can be involved, such as the hormone receptor coactivator ACTR, which is itself a HAT that acts on H3 and H4. One explanation for the presence of multiple HAT activities in a coactivating complex is that each HAT has a different specificity, and that multiple, different acetylation events are required for activation. This enables the picture for the action of coactivators to be redrawn, as shown in FIGURE 1 , where RNA polymerase II is bound at a hypersensitive site and coactivators are acetylating histones in the nucleosomes in the vicinity.
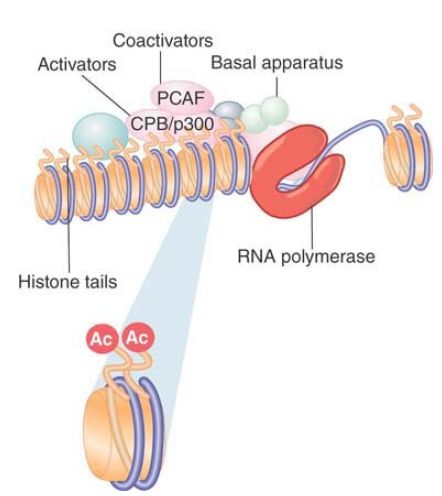
FIGURE 1. Coactivators may have HAT activities that acetylate the tails of nucleosomal histones.
Group A HATs, like ATP-dependent remodeling enzymes, are typically found in large complexes. FIGURE 2 shows a simplified model for their behavior. HAT complexes can be targeted to DNA by interactions with DNA-binding factors. The complex also contains effector subunits that affect chromatin structure or act directly on transcription. It is likely that at least some of the effectors require the acetylation event in order to act (such as the deubiquitylation activity of SAGA).

FIGURE 2 . Complexes that control acetylation levels have targeting subunits that determine their sites of action (usually subunits that interact with site-specific DNA-binding proteins), HAT or HDAC enzymes that acetylate or deacetylate histones, and effector subunits that have other actions on chromatin or DNA.
The effect of acetylation may be both quantitative and qualitative. In cases where the effect of charge neutralization on chromatin structure is key, a certain minimal number of acetyl groups should be required to have an effect, and the exact positions at which they occur are largely irrelevant. In the case where the role of acetylation is primarily in the creation of a binding site (for a bromodomain-containing factor, for example), the specific position of the acetylation event will be critical. The existence of complexes containing multiple HAT activities might be interpreted either way—if individual enzymes have different specificities, multiple activities might be needed either to acetylate a sufficient number of different positions or because the individual events are necessary for different effects upon transcription. At replication, it appears (at least with respect to histone H4) that acetylation at any two of three particular positions is adequate, favoring a quantitative model in this case. Where chromatin structure is changed to affect transcription, acetylation at specific positions is important .
As acetylation is linked to activation, deacetylation is linked to transcriptional repression. Whereas site-specific activators recruit coactivators with HAT activity, site-specific repressor proteins can recruit corepressor complexes, which often contain HDAC activity. In yeast, mutations in SIN3 and RPD3 result in increased expression of a variety of genes, indicating that Sin3 and Rpd3 proteins act as repressors of transcription. Sin3 and Rpd3 are recruited to a number of genes by interacting with the DNA-binding protein Ume6, which binds to the URS1 (upstream repressive sequence) element. The complex represses transcription at the promoters containing URS1, as illustrated in FIGURE 3. Rpd3 is a histone deacetylase, and its recruitment leads to deacetylation of nucleosomes at the promoter. Rpd3 and its homologs are present in multiple HDAC complexes found in eukaryotes from yeast to humans; these large complexes are typically built around Sin3 and its homologs.

FIGURE 3. A repressor complex contains three components: a DNA-binding subunit, a corepressor, and a histone deacetylase.
In mammalian cells, Sin3 is part of a repressive complex that includes histone-binding proteins and the Rpd3 homologs HDAC1 and HDAC2. This corepressor complex can be recruited by a variety of repressors to specific gene targets. The bHLH family of transcription regulators includes activators that function as heterodimers, including MyoD. This family also includes repressors, in particular the heterodimer Mad–Max, where Mad can be any one of a group of closely related proteins. The Mad–Max heterodimer (which binds to specific DNA sites) interacts with Sin3–HDAC1/2 complex and requires the deacetylase activity of this complex for repression. Similarly, the SMRT corepressor (which enables retinoid hormone receptors to repress certain target genes) binds mSin3, which, in turn, brings the HDAC activities to the site.
Another means of bringing HDAC activities to a DNA site can be an interaction with MeCP2, a protein that binds to methylated cytosines, a mark of transcriptional silencing . Absence of histone acetylation is also a feature of heterochromatin. This is true of both constitutive heterochromatin (typically involving regions of centromeres or telomeres) and facultative heterochromatin (regions that are inactivated in one cell although they may be active in another). Typically the N-terminal tails of histones H3 and H4 are not acetylated in heterochromatic regions .
 الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
 اخر الاخبار
اخر الاخبار
اخبار العتبة العباسية المقدسة

الآخبار الصحية















 قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة
قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة "المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة
"المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة (نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)
(نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)


















