

النبات

مواضيع عامة في علم النبات

الجذور - السيقان - الأوراق

النباتات الوعائية واللاوعائية

البذور (مغطاة البذور - عاريات البذور)

الطحالب

النباتات الطبية


الحيوان

مواضيع عامة في علم الحيوان

علم التشريح

التنوع الإحيائي

البايلوجيا الخلوية


الأحياء المجهرية

البكتيريا

الفطريات

الطفيليات

الفايروسات


علم الأمراض

الاورام

الامراض الوراثية

الامراض المناعية

الامراض المدارية

اضطرابات الدورة الدموية

مواضيع عامة في علم الامراض

الحشرات


التقانة الإحيائية

مواضيع عامة في التقانة الإحيائية


التقنية الحيوية المكروبية

التقنية الحيوية والميكروبات

الفعاليات الحيوية

وراثة الاحياء المجهرية

تصنيف الاحياء المجهرية

الاحياء المجهرية في الطبيعة

أيض الاجهاد

التقنية الحيوية والبيئة

التقنية الحيوية والطب

التقنية الحيوية والزراعة

التقنية الحيوية والصناعة

التقنية الحيوية والطاقة

البحار والطحالب الصغيرة

عزل البروتين

هندسة الجينات


التقنية الحياتية النانوية

مفاهيم التقنية الحيوية النانوية

التراكيب النانوية والمجاهر المستخدمة في رؤيتها

تصنيع وتخليق المواد النانوية

تطبيقات التقنية النانوية والحيوية النانوية

الرقائق والمتحسسات الحيوية

المصفوفات المجهرية وحاسوب الدنا

اللقاحات

البيئة والتلوث


علم الأجنة

اعضاء التكاثر وتشكل الاعراس

الاخصاب

التشطر

العصيبة وتشكل الجسيدات

تشكل اللواحق الجنينية

تكون المعيدة وظهور الطبقات الجنينية

مقدمة لعلم الاجنة


الأحياء الجزيئي

مواضيع عامة في الاحياء الجزيئي


علم وظائف الأعضاء


الغدد

مواضيع عامة في الغدد

الغدد الصم و هرموناتها

الجسم تحت السريري

الغدة النخامية

الغدة الكظرية

الغدة التناسلية

الغدة الدرقية والجار الدرقية

الغدة البنكرياسية

الغدة الصنوبرية

مواضيع عامة في علم وظائف الاعضاء

الخلية الحيوانية

الجهاز العصبي

أعضاء الحس

الجهاز العضلي

السوائل الجسمية

الجهاز الدوري والليمف

الجهاز التنفسي

الجهاز الهضمي

الجهاز البولي


المضادات الميكروبية

مواضيع عامة في المضادات الميكروبية

مضادات البكتيريا

مضادات الفطريات

مضادات الطفيليات

مضادات الفايروسات

علم الخلية

الوراثة

الأحياء العامة

المناعة

التحليلات المرضية

الكيمياء الحيوية

مواضيع متنوعة أخرى

الانزيمات
Bacteria Contain Regulator RNAs
المؤلف:
JOCELYN E. KREBS, ELLIOTT S. GOLDSTEIN and STEPHEN T. KILPATRICK
المصدر:
LEWIN’S GENES XII
الجزء والصفحة:
17-6-2021
2956
Bacteria Contain Regulator RNAs
KEY CONCEPTS
- Bacterial regulator RNAs are called small RNAs (sRNAs).
- Numerous sRNAs are bound by the protein Hfq, which increases their effectiveness.
- The oxyS sRNA activates or represses expression of approximately 40 loci at the posttranscriptional level.
- Tandem repeats can be transcribed into powerful antiviral RNAs called the CRISPR/Cas system.
Bacteria contain many—up to hundreds—of genes that encode regulator RNAs. These are short RNA molecules, ranging from about 50 to 200 nucleotides, that are collectively known as small RNAs, or sRNAs. Some of the sRNAs are general regulators that affect many target genes; others are specific for a single transcript. These sRNAs typically function as imperfect antisense RNAs; that is, their sequences are complementary to their target RNAs.
At what level does the antisense RNA inhibit expression? As with eukaryotic antisense RNAs, prokaryotic sRNAs could, in principle: (1) prevent transcription of the gene, (2) affect processing of its RNA product, (3) affect the translation of the messenger, or (4) affect the stability of the RNA. The action of sRNAs is primarily mediated by the formation of RNA–RNA duplex molecules.
Oxidative stress in Escherichia coli provides an interesting example of a general control system in which an sRNA is the regulator. When exposed to reactive oxygen species, bacteria respond by inducing antioxidant defense genes. Hydrogen peroxide activates the transcription activator OxyR, which controls the expression of several inducible genes. One of these genes is oxyS, which codes for an sRNA.
FIGURE 1 shows two salient features of the control of oxyS expression. In a wild-type bacterium under normal conditions, it is not expressed. The pair of gels on the left side of the figure shows that it is expressed at high levels in a mutant bacterium with a constitutively active oxyR gene. This identifies oxyS as a target for activation by oxyR. The pair of gels on the right side of the figure shows that oxyS RNA is transcribed within 1 minute of exposure to hydrogen peroxide.

FIGURE 1. The gels on the left show that oxyS RNA is induced in an oxyR constitutive mutant. The gels on the right show that oxyS RNA is induced within 1 minute of adding hydrogen peroxide to a wild-type culture.
Reprinted from Cell 90, S. Altuvia, et al., A small stable RNA …, pp. 43–53. Copyright 1997,
with permission from Elsevier
[http://www.sciencedirect.com/science/journal/00928674]. Photo courtesy of Gisela Storz, National Institutes of Health.
The oxyS RNA is a short sequence (109 nucleotides) that does not code for protein. It is a trans-acting antisense regulator that affects gene expression at the level of translation. It has about 40 target mRNAs; at some of them, it activates expression, and at others it represses expression. FIGURE 2 shows the mechanism of repression of one target, the flhA mRNA. Three stem-loop doublestranded RNA structures protrude in the secondary structure of oxyS RNA, and the loop closest to the 3′ terminus is complementary to a sequence just preceding the initiation codon of flhA mRNA. Base pairing between oxyS RNA and flhA RNA prevents the ribosome from binding to the initiation codon and therefore represses translation. A second pairing interaction involves a sequence within the coding region of the flhA mRNA.
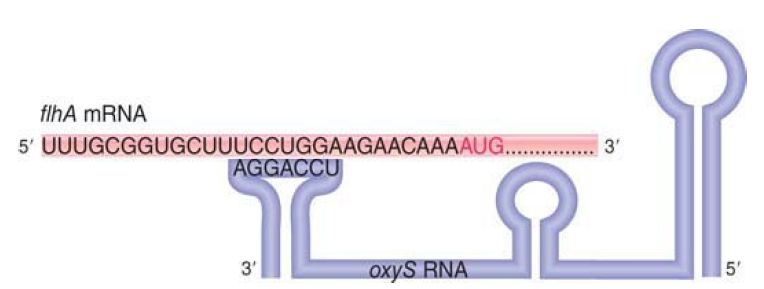
FIGURE 2. oxyS RNA inhibits translation of flhA mRNA by base pairing with a sequence just upstream of the AUG initiation codon.
Another target for oxyS is rpoS, the gene encoding an alternative sigma factor (which activates a general stress response). rpoS mRNA is negatively autoregulated by a stem-loop in the 5′ region of the message, which prevents ribosome access to the open reading frame (ORF). By reinforcing this, and thus inhibiting production of the sigma factor, oxyS ensures that the specific response to oxidative stress does not trigger the response that is appropriate for other stress conditions. The rpoS gene is also positively regulated by three other sRNAs (dsrA, arcZ, and rprA), which activate it by binding to the stem-loop region, opening it up, and making the ORF available to the ribosome. These four sRNAs appear to be global regulators that coordinate responses to various environmental conditions.
The actions of three of these sRNAs are assisted by an RNAbinding protein called Hfq (DsrA can act partly independently of Hfq) that acts to stabilize the sRNA–mRNA binding. The Hfq protein was originally identified as a bacterial host factor needed for replication of the RNA bacteriophage Qβ. It is related to the Sm proteins of eukaryotes that bind to many of the small nuclear RNAs (snRNAs) that have regulatory roles in gene expression . Mutations in its gene have many effects; this identifies it as a pleiotropic protein. Hfq binds to many of the sRNAs of E. coli, and it increases the effectiveness of oxyS RNA by enhancing its ability to bind to its target mRNAs. The effect of Hfq is probably mediated by causing a small change in the secondary structure of oxyS RNA that improves the exposure of the single-stranded sequences that pair with the target mRNAs. The vast potential that small RNAs possess in controlling so much of the life cycle of an organism is just beginning to be realized. A system of bacterial defense against foreign invaders, both viruses and certain plasmids, in the very well-known bacterium E. coli provides an example of just how much there is to learn about small RNAs. This adaptive immune system is based upon clusters of short palindromic repeats called CRISPRs (clusters of regularly interspersed short palindromic repeats) separated by hypervariable spacer sequences derived from captured phage and plasmids. These are widespread in both eubacteria and archaea. These hypervariable CRISPR spacer sequences are used to provide the host bacteria with resistance to further phage and plasmid infection, as shown in FIGURE 3.
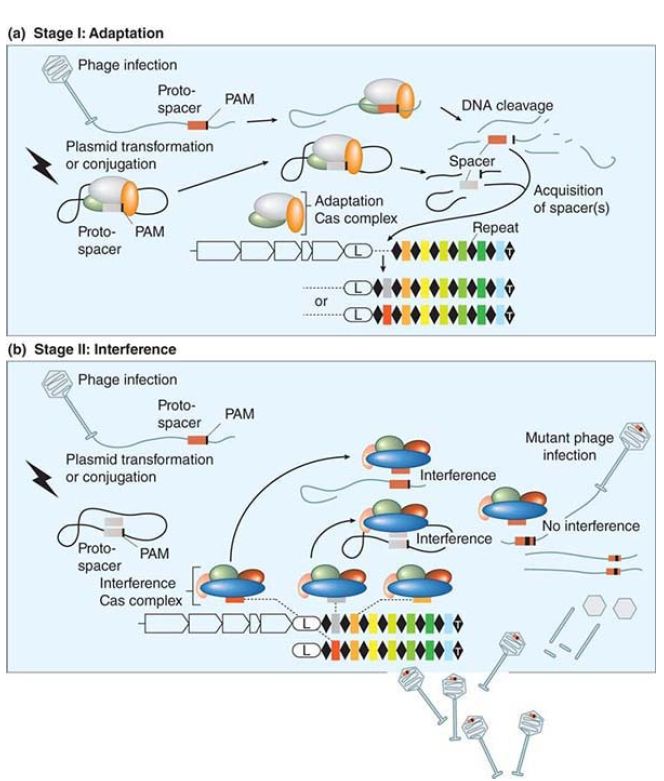
FIGURE 3. Adaptation and interference stages of the CRISPR/Cas system. (a) Stage I: Adaptation. Entry of foreign DNA into a cell through transformation, conjugation, or transduction can lead to acquisition of new DNA spacer(s) by the adaptation Cas complex (unknown protein assembly). If no spacer is acquired, the phage lytic cycle or plasmid replication can proceed (not shown).(b) Stage II: Interference. The interfering Cas complexes are bound to a crRNA produced from the transcription of the CRISPR locus and subsequent processing. A cell carrying a crRNA targeting a region (by perfect pairing) of foreign nucleic acid can interfere with the invasive genetic material and destroy it via an interference Cas complex (unknown protein assembly except for Cascade in E. coli). If there is no perfect pairing between the spacer and the protospacer (as in the case of a phage mutant), the CRISPR/Cas system is counteracted and replication of the invasive genetic material can occur.
Reproduced from H. Deveau, et al. Annu. Rev. Microbiol 64 (2010): 475–493.
The CRISPR defense system requires transcription of the repeatspacer array from a leader sequence (acting as a promoter) and is used in conjunction with an RNA-processing system of eight genes, called cas (CRISPR-associated) genes in E. coli K12, usually located adjacent to each CRISPR locus. These genes code for a variety of polymerases, nucleases (both DNA and RNA), helicases, and RNA-binding proteins. A multimeric complex of five Cas proteins can be identified and is called Cascade (CRISPRassociated complex for antiviral defense). The Cas complex is responsible not only for the interference stage but also for the adaptation stage, which processes the foreign invader for incorporation into the CRISPR locus. Three major families of CRISPR/Cas genes have been identified, depending on the specific Cas proteins in the genome.
The CRISPR region is transcribed into a long RNA, pre-crRNA, which is processed into short CRISPR RNAs of about 57 nucleotides containing a spacer flanked by two conserved partial repeats, the protospacer-adjacent motifs (PAMs). The model proposed is that these spacer/PAM RNAs, complementary to phage DNA protospacer sequences, are subsequently used as guides for the Cas interference machinery. Pairing is initiated by a high-affinity seed sequence at either end of the crRNA spacer sequence (similar to that seen in eukaryotic miRNA function, as described in the section later in this chapter, How Does RNA Interference Work?). The complex base pairs with the virus genome (or its RNA) to prevent expression of the phage genes and ultimately leads to degradation. Mutations in either the spacer DNA core seed sequence or the PAM sequence abolish CRISPR/Cas immunity by altering binding. The CRISPR/Cas system has been adapted for genome editing due to the precision with which a precisely targeted sequence can be altered in a genome .
These mechanisms offer powerful approaches for turning off genes at will and altering gene expression. It is not, however, necessarily a one-way street where a regulatory RNA is produced and simply turns off expression of a message. This system can also be balanced by the production of a counter protein that can bind to and interfere with the sRNA. Thus, dynamic systems can exist that can change over time according to demands placed on the cell.
The function of a regulatory gene can be investigated by introducing an antisense version. An extension of this technique is to place the antisense gene under the control of a promoter that is
itself subject to regulation. The target gene can then be turned off and on by regulating the production of antisense RNA. This technique allows investigation of the importance of the timing of expression of the target gene.
 الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
 اخر الاخبار
اخر الاخبار
اخبار العتبة العباسية المقدسة

الآخبار الصحية















 قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة
قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة "المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة
"المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة (نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)
(نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)


















