

النبات

مواضيع عامة في علم النبات

الجذور - السيقان - الأوراق

النباتات الوعائية واللاوعائية

البذور (مغطاة البذور - عاريات البذور)

الطحالب

النباتات الطبية


الحيوان

مواضيع عامة في علم الحيوان

علم التشريح

التنوع الإحيائي

البايلوجيا الخلوية


الأحياء المجهرية

البكتيريا

الفطريات

الطفيليات

الفايروسات


علم الأمراض

الاورام

الامراض الوراثية

الامراض المناعية

الامراض المدارية

اضطرابات الدورة الدموية

مواضيع عامة في علم الامراض

الحشرات


التقانة الإحيائية

مواضيع عامة في التقانة الإحيائية


التقنية الحيوية المكروبية

التقنية الحيوية والميكروبات

الفعاليات الحيوية

وراثة الاحياء المجهرية

تصنيف الاحياء المجهرية

الاحياء المجهرية في الطبيعة

أيض الاجهاد

التقنية الحيوية والبيئة

التقنية الحيوية والطب

التقنية الحيوية والزراعة

التقنية الحيوية والصناعة

التقنية الحيوية والطاقة

البحار والطحالب الصغيرة

عزل البروتين

هندسة الجينات


التقنية الحياتية النانوية

مفاهيم التقنية الحيوية النانوية

التراكيب النانوية والمجاهر المستخدمة في رؤيتها

تصنيع وتخليق المواد النانوية

تطبيقات التقنية النانوية والحيوية النانوية

الرقائق والمتحسسات الحيوية

المصفوفات المجهرية وحاسوب الدنا

اللقاحات

البيئة والتلوث


علم الأجنة

اعضاء التكاثر وتشكل الاعراس

الاخصاب

التشطر

العصيبة وتشكل الجسيدات

تشكل اللواحق الجنينية

تكون المعيدة وظهور الطبقات الجنينية

مقدمة لعلم الاجنة


الأحياء الجزيئي

مواضيع عامة في الاحياء الجزيئي


علم وظائف الأعضاء


الغدد

مواضيع عامة في الغدد

الغدد الصم و هرموناتها

الجسم تحت السريري

الغدة النخامية

الغدة الكظرية

الغدة التناسلية

الغدة الدرقية والجار الدرقية

الغدة البنكرياسية

الغدة الصنوبرية

مواضيع عامة في علم وظائف الاعضاء

الخلية الحيوانية

الجهاز العصبي

أعضاء الحس

الجهاز العضلي

السوائل الجسمية

الجهاز الدوري والليمف

الجهاز التنفسي

الجهاز الهضمي

الجهاز البولي


المضادات الميكروبية

مواضيع عامة في المضادات الميكروبية

مضادات البكتيريا

مضادات الفطريات

مضادات الطفيليات

مضادات الفايروسات

علم الخلية

الوراثة

الأحياء العامة

المناعة

التحليلات المرضية

الكيمياء الحيوية

مواضيع متنوعة أخرى

الانزيمات
Supercoiling Affects the Structure of DNA
المؤلف:
JOCELYN E. KREBS, ELLIOTT S. GOLDSTEIN and STEPHEN T. KILPATRICK
المصدر:
LEWIN’S GENES XII
الجزء والصفحة:
23-2-2021
4131
Supercoiling Affects the Structure of DNA
KEY CONCEPTS
-Supercoiling occurs only in “closed” DNA with no free ends.
-Closed DNA is either circular DNA or linear DNA in which the ends are anchored so that they are not free to rotate.
-A closed DNA molecule has a linking number (L), which is the sum of twist (T) and writhe (W).
-The linking number can be changed only by breaking and reforming bonds in the DNA backbone.
The two strands of DNA are wound around each other to form a double helical structure (described in detail in the next section); the double helix can also wind around itself to change the overall conformation, or topology, of the DNA molecule in space. This is called supercoiling. The effect can be imagined like a rubber band twisted around itself. Supercoiling creates tension in the DNA; thus, it can occur only if the DNA has no free ends (otherwise the free ends can rotate to relieve the tension) or in linear DNA (FIGURE 1, top) if it is anchored to a protein scaffold, as in eukaryotic chromosomes. The simplest example of a DNA with no free ends is a circular molecule. The effect of supercoiling can be seen by comparing the nonsupercoiled circular DNA lying flat in Figure 1 (center) with the supercoiled circular molecule that forms a twisted, and therefore more condensed, shape (Figure 1, bottom).
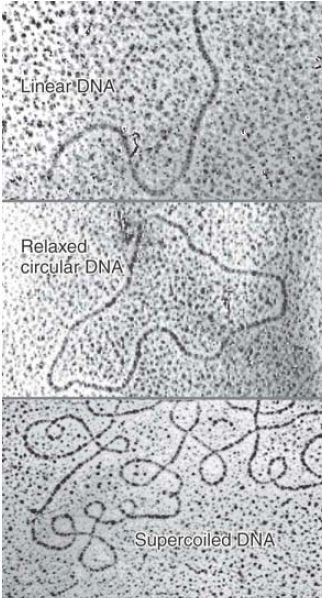
FIGURE 1. Linear DNA is extended (top); a circular DNA remains extended if it is relaxed (nonsupercoiled; center); but a supercoiled DNA has a twisted and condensed form (bottom). Photos courtesy of Nirupam Roy Choudhury, International Centre for Genetic Engineering and Biotechnology (ICGEB).
The consequences of supercoiling depend on whether the DNA is twisted around itself in the same direction as the two strands within the double helix (clockwise) or in the opposite direction. Twisting in the same direction produces positive supercoiling, which overwinds the DNA so that there are fewer base pairs per turn.
Twisting in the opposite direction produces negative supercoiling, or underwinding, so there are more base pairs per turn. Both types of supercoiling of the double helix in space are tensions in the DNA (which is why DNA molecules with no supercoiling are said to be “relaxed”). Negative supercoiling can be thought of as creating tension in the DNA that is relieved by the unwinding of the double helix. The effect of severe negative supercoiling is to generate a region in which the two strands of DNA have separated (technically, zero base pairs per turn).
Topological manipulation of DNA is a central aspect of all of its functional activities (e.g., recombination, replication, and transcription) as well as of the organization of its higher order structure. All synthetic activities involving double-stranded DNA require the strands to separate. The strands do not simply lie side by side though; they are intertwined. Their separation therefore requires the strands to rotate about each other in space. Some possibilities for the unwinding reaction are illustrated in FIGURE 2.
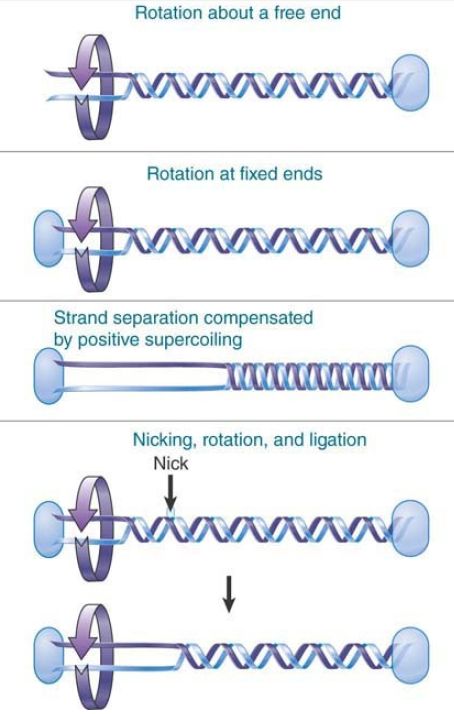
FIGURE 2. Separation of the strands of a DNA double helix can be achieved in several ways.
Unwinding a short linear DNA presents no problems, because the DNA ends are free to spin around the axis of the double helix to relieve any tension. DNA in a typical chromosome, however, is not only extremely long but also coated with proteins that serve to anchor the DNA at numerous points. As a result, even a linear eukaryotic chromosome does not functionally possess free ends.
Consider the effects of separating the two strands in a molecule whose ends are not free to rotate. When two intertwined strands are pulled apart from one end, the result is to increase their winding about each other farther along the molecule, resulting in positive supercoiling elsewhere in the molecule to balance the underwinding generated in the single-stranded region. The problem can be overcome by introducing a transient nick in one strand. An internal free end allows the nicked strand to rotate about the intact strand, after which the nick can be sealed. Each repetition of the nicking and sealing reaction releases one superhelical turn.
A closed molecule of DNA can be characterized by its linking number (L), which is the number of times one strand crosses over the other in space. Closed DNA molecules of identical sequence can have different linking numbers, reflecting different degrees of supercoiling. Molecules of DNA that are the same except for their linking numbers are called topological isomers.
The linking number is made up of two components: the writhing number (W) and the twisting number (T). The twisting number, T, is a property of the double helical structure itself, representing the rotation of one strand about the other. It represents the total number of turns of the duplex and is determined by the number of base pairs per turn. For a relaxed closed circular DNA lying flat in a plane, T is the total number of base pairs divided by the number of base pairs per turn. The writhing number, W, represents the turning of the axis of the duplex in space. It corresponds to the intuitive concept of supercoiling but does not have exactly the same quantitative definition or measurement. For a relaxed molecule, W= 0, and the linking number equals the twist.
We are often concerned with the change in linking number, ΔL, given by the equation:
ΔL = ΔW + ΔT
The equation states that any change in the total number of revolutions of one DNA strand about the other can be expressed as the sum of the changes of the coiling of the duplex axis in space
(ΔW) and changes in the helical repeat of the double helix itself (ΔT). In the absence of protein binding or other constraints, the twist of DNA does not tend to vary—in other words, the 10.5 base pairs per turn (bp/turn) helical repeat is a very stable conformation for DNA in solution. Thus, any ΔL is mostly likely to be expressed by a change in W; that is, by a change in supercoiling.
A decrease in linking number (that is, a change of −ΔL) corresponds to the introduction of some combination of negative supercoiling (ΔW) and/or underwinding (ΔT). An increase in linking number, measured as a change of +ΔL, corresponds to an increase in positive supercoiling and/or overwinding.
We can describe the change in state of any DNA by the specific linking difference, σ = ΔL/L0, for which L0 is the linking number when the DNA is relaxed. If all of the change in the linking number is due to change in W (that is, ΔT = 0), the specific linking difference equals the supercoiling density. In effect, σ, as defined in terms of ΔL/L0, can be assumed to correspond to supercoiling density so long as the structure of the double helix itself remains constant.
The critical feature about the use of the linking number is that this parameter is an invariant property of any individual closed DNA molecule. The linking number cannot be changed by any deformation short of one that involves the breaking and rejoining of strands. A circular molecule with a particular linking number can express the number in terms of different combinations of T and W, but it cannot change their sum so long as the strands are unbroken.
(In fact, the partitioning of L between T and W prevents the assignment of fixed values for the latter parameters for a DNA molecule in solution.)
The linking number is related to the actual enzymatic events by which changes are made in the topology of DNA. The linking number of a particular closed molecule can be changed only by
breaking one or both strands, using the free end to rotate one strand about the other, and rejoining the broken ends. When an enzyme performs such an action, it must change the linking number by an integer; this value can be determined as a characteristic of the reaction. The reactions to control supercoiling in the cell are performed by topoisomerase enzymes.
 الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
 اخر الاخبار
اخر الاخبار
اخبار العتبة العباسية المقدسة

الآخبار الصحية















 قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة
قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة "المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة
"المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة (نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)
(نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)


















