


 النبات
النبات
 الحيوان
الحيوان
 الأحياء المجهرية
الأحياء المجهرية
 علم الأمراض
علم الأمراض
 التقانة الإحيائية
التقانة الإحيائية
 التقنية الحيوية المكروبية
التقنية الحيوية المكروبية
 التقنية الحياتية النانوية
التقنية الحياتية النانوية
 علم الأجنة
علم الأجنة
 الأحياء الجزيئي
الأحياء الجزيئي
 علم وظائف الأعضاء
علم وظائف الأعضاء
 الغدد
الغدد
 المضادات الحيوية
المضادات الحيوية|
Read More
Date: 16-6-2021
Date: 19-12-2015
Date: 15-5-2016
|
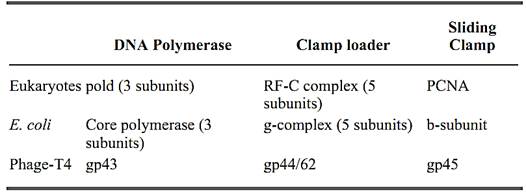
Clamp Loaders, Processivity Complex
Chromosomal replicases, the DNA polymerases that replicate chromosomes, are multiprotein complexes characterized by the high processivity of their DNA synthesis. These replicating machines polymerize thousands of nucleotides without dissociating from the template. The replicases of the three well-characterized systems (Escherichia coli, eukaryotes, and bacteriophage T4), which span the evolutionary spectrum, are similar in function, structure, and overall organization. Their remarkable processivity is achieved by a ring-shaped processivity factor (or “sliding clamp”) that binds the polymerase catalytic unit and tethers it to the DNA. Another complex of accessory proteins is required to load the clamp onto the DNA. The sliding clamp does not have any affinity for DNA and, therefore, other proteins are needed to assemble the clamp around the DNA. The accessory protein complex (“clamp loader”; also called a “molecular matchmaker”) binds to the primer terminus and couples ATP hydrolysis to the assembly of the ring around the DNA primer. Therefore, the replicase can be thought of as having three components: (1) a polymerase, (2) a clamp loader complex that assembles the clamp around DNA and (3) a DNA sliding clamp (Table 1).
Table 1. Three-Component Structures of Chromosomal Replicases
1. The Clamp Loaders
The three best-studied replicases are the E. coli DNA polymerase III holoenzyme (polIII), the eukaryotic polymerase d (pold), and the replicase of phage T4. These three multisubunit complexes share structural and functional similarities (reviewed in Refs. 1-3). The clamp loaders for these systems are the g-complex of prokaryotes, the replication factor-C (RF-C) complex (also called Activator-1) of eukaryotes, and a complex of the products of gene 44 and gene 62 (gp44/62) of phage T4. The g-complex is composed of five subunits called g, d, d′, c and y. RF-C is also a five-subunit complex, composed of one large and four small subunits (p140, p40, p38, p37, and p36). gp44/62 contains only two polypeptides gp44 and gp62. In the complex, a tetramer of gp44 is tightly associated with one gp62. Interestingly, the three clamp loaders share amino acid similarities among the subunits (4, 5.(
A detailed mechanism by which these clamp loaders operate in assembling the sliding clamp around DNA is not yet known. The basic steps of the loading process, however, have been elucidated (Fig1.). In general, the clamp loader recognizes the 3′ end of the junction between the single strand and duplex DNA (primer/template) and uses ATP hydrolysis to assemble the clamp around the primer. The status of the clamp loader after assembling the clamp around DNA is not yet clear. It has not yet been determined whether the clamp loader leaves the clamp on the DNA, where it interacts with the polymerase to initiate processive DNA synthesis, or if the clamp loader is also needed to assist binding the polymerase to the clamp (the different features of each clamp loader are discussed later).
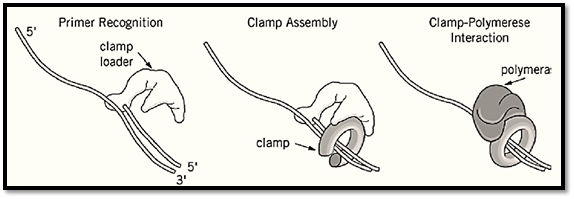
Figure 1. A model for clamp loader activity. The clamp loader recognizes the 3′ terminus of a primer and assembles the sliding clamp around the DNA. Following assembly, the clamp interacts with the polymerase and tethers it to the DNA f processive DNA synthesis.
In prokaryotes and eukaryotes, the clamp loader also functions as a clamp unloader (6). Upon completion of an Okazaki fragment, the polymerase rapidly dissociates from the clamp (7-9), leaving the clamp on the DNA (10). The clamps of polIII (the b subunit) and pold [proliferating cell nuclear antigen (PCNA)] are relatively stable on DNA (6, and references therein). Therefore, it was postulated that the sliding clamp remains assembled around the DNA when the synthesis of an Okazaki fragment is complete. During lagging strand synthesis, a new sliding clamp is needed for the synthesis of each Okazaki fragment. In E. coli about 10 times more Okazaki fragments are formed during replication than there are b-subunits within the cell. In human cells, the number of Okazaki fragments formed during one round of chromosomal replication has been estimated to be 100 times greater than the amount of PCNA. Therefore, recycling of the clamp (b and PCNA) is needed to fulfill the requirement for a constant supply of clamps for further DNA synthesis. In phage T4, the clamp readily dissociates from DNA upon completion of an Okazaki fragment. Thus, the clamp loader has a dual function during DNA replication: (1) loading of the sliding clamp onto DNA to initiate processive DNA synthesis and (2) recycling the clamps on the lagging strand.
1.1. g-Complex
The g-complex is composed of five subunits (g, d, d′, c, y) (reviewed in Ref. 11). The genes encoding them have been identified, and the purified proteins have been used to study the function of the individual subunits and subassemblies of the g-complex (discussed later). Four of the subunits (d, d′, c and y) are encoded by unique genes. The g-subunit is formed from the same gene (dnaX) that encodes t (another subunit of polIII) by an efficient translational frameshift mechanism that produces g in an amount equal to t (reviewed in Ref. 12). As a result, the g-subunit consists of the N-terminal 430 residues of t followed by a unique C-terminal Glu residue. In addition, the g-and d′-subunits show amino acid sequence similarity.
No one subunit alone can assemble the b sliding clamp around DNA (11 and references therein). At low ionic strength, a combination of g and d assemble b onto DNA, but the reaction is feeble. The d′-, c- and y-subunits are needed for an efficient loading reaction under physiological conditions (13).
The g-complex has only weak affinity for single-stranded and duplex DNA, and it exhibits weak DNA-dependent ATPase activity, but the ATPase activity is stimulated by the sliding clamp (b-subunit) (14). The best DNA effector for the ATPase activity is a singly primed template, indicating an interaction between the g-complex and the primer/template junction. The g-subunit is the only subunit of the g-complex with an ATP-binding motif, and it binds ATP (15). Furthermore, mutation of the ATP-binding site of g within the g-complex destroys the ATPase activity and the ability to assemble the b-ring onto DNA. The g-subunit, however, lacks significant ATP hydrolysis activity even in the presence of DNA (14, 15), and the d- and d′-subunits are required for ATPase activity, implying that the gdd′ complex recognizes DNA. The g-complex binds b in the presence of ATP. The d subunit interacts with b, with a strength similar to that of the entire g-complex (16), indicating that b binds the g-complex mainly through the d-subunit. Interestingly, however, the interaction between the b-subunit and d does not depend on ATP (whereas the g-complex requires ATP to bind the clamp). These apparently contradictory observations can be explained if the d subunit is buried within the g-complex and ATP induces conformational changes that lead to the interaction of d with b (16). The g-complex also interacts with single-stranded DNA binding protein (SSB) via the c subunit (11). These interactions are important for efficient clamp assembly at physiological ionic strength.
The results obtained with the individual subunits and their subassemblies demonstrate the basic features of the g-complex action during loading of the clamp onto DNA (Fig. 1). Upon binding of ATP, the g-complex undergoes conformational changes leading to interaction of the d-subunit with b. The g-complex recognizes a primed template, aided by the interaction between c and SSB. Assembly of the ring around the DNA primer stimulates the ATPase activity of the g-complex. ATP hydrolysis results in conformational changes such that d is again buried within the g-complex leaving the clamp around the DNA primer. Now the clamp interacts with the polymerase to initiate processive DNA synthesis (Fig. 1).
1.2. Replication Factor-C (RF-C(
RF-C was first isolated from human 293 cells as an essential replication factor for the in vitro replication of simian virus-40 (SV-40) (17). The factor was purified later from different sources, including yeast and human cell lines (18 and references therein), based on its ability to stimulate processive DNA synthesis by pold in the presence of PCNA and ATP.
Purified RF-C is a five-subunit complex (p140, p40, p38, p37, and p36). The genes that encode all RF-C subunits from yeast and humans have been identified (4, 5 and references therein), and all are essential in yeast. Although encoded by different genes, they show extensive amino acid sequence similarities in the central region of the protein (reviewed in Ref. 5).
Early biochemical studies determined the role of RF-C as a clamp loader of PCNA and suggested a mode of action during the assembly reaction (Fig. 1). RF-C binds preferentially to single-strand/duplex DNA (template/primer) junction in the presence of ATP (19, 20). PCNA interacts with the RF-C/DNA complex. The DNA-dependent ATPase activity of RF-C is stimulated by the binding of PCNA. The hydrolysis of ATP leads to conformational changes within the RF-C, which locks the PCNA sliding clamp around the DNA. Then the clamp interacts with the polymerase to initiate processive DNA synthesis (Fig. 1).
Biochemical analysis of RF-C individual subunits and the protein–protein interactions within the RF-C complex is underway (eg, 21, 22). Purified proteins, subcomplexes and mutational analysis have shed light on the function of the individual subunits and subassemblies of the RF-C complex. Although each subunit contains an ATP-binding motif, only p40 binds ATP (20). Yeast p36 (23) and a subcomplex of human RF-C (p36-p37-p40) exhibit DNA-dependent ATPase activity. The DNA-binding activity resides within the large subunit (p140) (20), although the p37 subunit also binds DNA weakly. All of the subunits, except p37, bind PCNA (24-26). The intact complex, however, is needed for the assembly of PCNA around the DNA and, to date, no subunit combination analyzed performs this task. Unloading is probably a simpler mechanism, as the p40 subunit is sufficient to unload PCNA from DNA and also interacts with pold (26). The role of this interaction in DNA synthesis, however, is not yet clear. RF-C also interacts with SSB. The interaction between RF-C and SSB might be needed for the assembly of the clamp, as demonstrated for the E. coli g-complex.
1.3. gp44/62
Similar to the g-complex and RF-C, the clamp loader of phage T4, the gp44/62 complex, is also composed of five subunits (Table 1). In this case, however, only two polypeptides form the pentamer, where a tetramer of gp44 binds a single subunit of gp62 (27). Like the clamp loader of eukaryotes and prokaryotes, the gp44/62 complex exhibits ATPase activity that is stimulated by the sliding clamp (gp45) and by DNA (28, reviewed in Ref. 29). A DNA structure that resembles a template/primer junction is the best effector for stimulating ATPase activity (27). Cross-linking and protein-DNA footprinting assays demonstrate that the clamp loader interacts with DNA in the absence of gp45, but these interactions are relatively weak, and the clamp is needed for binding to DNA. Studies with individual subunits of the gp44/62 complex revealed that the gp44 exhibits the ATPase activity, whereas gp62 interacts with the clamp (31).
The exact mechanism of action of the gp44/62 complex is not yet fully understood. The overall features of its activity, however, have been determined and are similar to those of the g-complex and RF-C (Fig. 1). The gp44/62 complex interacts with gp45 in an ATP-dependent manner. The complex recognizes the primer terminus, which stimulates the ATPase activity of the clamp loader, bringing about conformational changes that lock the clamp around the primer and allow it to interact with the polymerase to initiate processive DNA synthesis.
2. Concluding Remarks
The polymerases responsible for replicating chromosomal DNA during cell division are multiprotein complexes. The processivity of these enzymes relies on a ring-shaped protein that encircles DNA and tethers the catalytic unit to DNA for processive DNA synthesis. The mechanisms by which the clamp loaders assemble the ring around DNA and disassemble them from DNA are only beginning to be understood. The complexity of this mechanism is inferred from the observation that it requires the coordinate activity of several polypeptides in eukaryotes, prokaryotes, and phage T4. Future studies are needed to elucidate the mechanisms by which the clamp loaders operate in the loading and unloading reactions. Purification of the clamp loader complexes and the individual subunits will enable further analysis of these mechanisms.
References
1. B. Stillman (1994) Cell 78, 725–728.
2. Z. Kelman and M. O''Donnell (1994) Curr. Opinion Genet. Dev. 4, 185–195.
3. B. Stillman (1996) DNA Replication in Eukaryotic Cells. M. L. DePamphilis, ed., CSH Laboratory Press, Cold Spring Harbor, NY, pp. 435–460.
4. M. O''Donnell, R. Onrust, F. B. Dean, M. Chen, and J. Hurwitz (1993) Nucleic Acids Res. 21, 3-1.
5. G. Cullmann, K. Fien, R. Kobayashi, and B. Stillman (1995) Mol. Cell. Biol. 15, 4661–4671.
6. Yao, J. Turner, Z. Kelman, P. T. Stukenberg, F. Dean, D. Shechter, Z.-Q. Pan, J. Hurwitz, and M. O''Donnell (1996) Genes to Cell 1, 101–113.
7. C. A. Wu, E. L. Zechner, and K. J. Marians (1992) J. Biol. Chem. 267, 4030–4044.
8. P. T. Stukenberg, J. Turner, and M. O''Donnell (1994) Cell 78, 877–887.
9. K. J. Hacker and B. M. Alberts (1994) J. Biol. Chem. 269, 24221–24228.
10. A. Yuzhakov, J. Turner, and M. O''Donnell (1996) Cell 86, 877–886.
11. Z. Kelman and M. O''Donnell (1995) Ann. Rev. Biochem. 64, 171–200.
12. C. S. McHenry (1988) Ann. Rev. Biochem. 57, 519–550.
13. M. O''Donnell and P. S. Studwell (1990) J. Biol. Chem. 265, 1179–1187.
14. R. Onrust, P. T. Stukenberg, and M. O''Donnell (1991) J. Biol. Chem. 266, 21681–21686.
15. Z. Tsuchihashi and A. Kornberg (1989) J. Biol. Chem. 264, 17790–17795.
16. V. Naktinis, R. Onrust, L. Fang, and M. O''Donnell (1995) J. Biol. Chem. 270, 13358–13365.
17. T. Tsurimoto and B. Stillman (1989) Mol. Cell. Biol. 9, 609–619.
18. U. Hubscher, G. Maga, and V. N. Podust (1996) DNA Replication in Eukaryotic Cells, M. L. DePamphilis (ed.), CSH Laboratory Press, Cold Spring Harbor, NY, pp. 525–543.
19. S.-H. Lee, A. D. Kwong, Z.-Q. Pan, and J. Hurwitz (1991) J. Biol. Chem. 266, 594–602.
20. T. Tsurimoto and B. Stillman (1991) J. Biol. Chem. 266, 1950–1960.
21. F. Uhlmann, J. Chi, H. Flores-Rozas, F. B. Dean, J. Finkelstein, M. O''Donnell, and J. Hurwitz (1996) Proc. Natl. Acad. Sci. USA 93, 6521–6526.
22. V. N. Podust and E. Fanning (1997) J. Biol. Chem. 272, 6303–6310.
23. X. Li and P. M. Burgers (1994) Proc. Natl. Acad. Sci. USA 91, 868–872.
24. M. A. McAlear, E. A. Howell, K. K. Espenshade, and C. Holm (1994) Mol. Cell. Biol. 14, 4390-4397.
25. Chen, Z.-Q. Pan, and J. Hurwitz (1992) Proc. Natl. Acad. Sci. USA 89, 2516–2520.
26. Z.-Q. Pan, M. Chen, and J. Hurwitz (1993) Proc. Natl. Acad. Sci. USA 90, 6–10.
27. T. C. Jarvis, L. S. Paul, and P. H. von Hipple (1989) J. Biol. Chem. 264, 12709–12716.
28. J. R. Piperno and B. M. Alberts (1978) J. Biol. Chem. 253, 5174–5179.
29. M. C. Young, M. K. Reddy, T. C. Jarvis, E. P. Gogol, M. K. Dolejsi, and P. H. von Hippel (1994) Molecular Biology of Bacteriophage T4. (J. D. Karam, ed.), American Society for Microbiology, Washington, DC, pp. 313–317.
30.N. G. Nossel (1994) Molecular Biology of Bacteriophage T4. (J. D. Karam, ed.), American Society for Microbiology, Washington, DC, pp. 43–53.
31. J. Rush, T.-C. Lin, M. Quinones, E. K. Spicer, I. Douglas, K. R. Williams, and W. H. Konigsberg (1989) J. Biol. Chem. 264, 10943–10953.



|
|
|
|
التوتر والسرطان.. علماء يحذرون من "صلة خطيرة"
|
|
|
|
|
|
|
مرآة السيارة: مدى دقة عكسها للصورة الصحيحة
|
|
|
|
|
|
|
نحو شراكة وطنية متكاملة.. الأمين العام للعتبة الحسينية يبحث مع وكيل وزارة الخارجية آفاق التعاون المؤسسي
|
|
|