


 النبات
النبات
 الحيوان
الحيوان
 الأحياء المجهرية
الأحياء المجهرية
 علم الأمراض
علم الأمراض
 التقانة الإحيائية
التقانة الإحيائية
 التقنية الحيوية المكروبية
التقنية الحيوية المكروبية
 التقنية الحياتية النانوية
التقنية الحياتية النانوية
 علم الأجنة
علم الأجنة
 الأحياء الجزيئي
الأحياء الجزيئي
 علم وظائف الأعضاء
علم وظائف الأعضاء
 الغدد
الغدد
 المضادات الحيوية
المضادات الحيوية|
Read More
Date: 22-5-2021
Date: 20-12-2015
Date: 10-5-2016
|
Antibody
Antibodies are specific proteins, termed immunoglobulins, that are produced by B cells upon stimulation with antigens, which may be proteins, polysaccharides, nucleic acids, and so on, either in soluble form or as part of complex cellular organisms, such as bacteria, parasites, viruses, eukaryotic cells of animal or plant tissues, and pollens. In fact, the immune system is so built as to have the potential to make antibodies against any macromolecule of the living world. It was reported at the end of the nineteenth century by von Behring and Kitasato that a group of guinea pigs immunized against a sublethal dose of diphtheria toxin became resistant to diphtheria, whereas another group that received tetanus toxin resisted further challenge with a normally lethal dose of tetanus toxin. This key experiment showed that the protection was specific toward the immunizing agent. Thus, one master word in immunology is specificity, and this immediately raises the problem of what immunologists call the repertoire. How is it possible to make the billions of different molecules that are potentially necessary to assume specific recognition of an obviously astronomical number of potential antigens? What structures are recognized on an antigen? What are the structural bases for antibody specificity? All these are basic questions that have been solved progressively during the past 40 years.
The protein nature of antibodies was established by M. Heidelberger in 1928, by showing that protein nitrogen was present in a specific antigen–antibody precipitate in which the antigen was a polysaccharide, which must have originated from the antibody part of the precipitate. Progress in the knowledge of the structure of antibodies paralleled the emergence of new technological tools in biochemistry and in molecular biology. Analytical ultracentrifugation performed by Kabat and Tiselius at the end of the 1930 indicated that antibodies were found in several classes of sizes, mostly with sedimentation coefficients of between 19S and 7S. Electrophoretic characterization of serum, also studied by Kabat in the same period, revealed that antibodies were located in the globulin part of the spectrum, mostly in the slowest moving g-globulin fraction. This name remained in use for years until it was replaced by a more generic one, that of immunoglobulins. Immunoglobulins are thus defined as the immune proteins synthesized specifically by B cells. They are expressed in two forms : (1) as integral membrane proteins at the surface of B lymphocytes, where they represent the B-cell receptor (BCR), by analogy with the T-cell receptor (TCR) expressed at the surface of T cells; and (2) as a soluble form, secreted in the bloodstream by the plasma cells, which represent the terminal stage of differentiation of the B lineage. This soluble form is what immunologists call circulating antibodies, or simply antibodies.
A main difficulty encountered in studying antibody structure was due to their extraordinary heterogeneity, which deserves some comments. As a rule, when an antigen such as hemoglobin or serum albumin, or in fact any protein, is used as an immunogen, a large array of antibodies with discrete fine specificities is synthesized. At the surface of a protein, many discrete regions are recognized by as many distinct antibodies. These regions are called antigenic determinants, or epitopes. For a protein of molecular weight 50 kDa, there are 10, 20, or more epitopes that could be identified by distinct antibodies. But antibody heterogeneity is not limited to the mosaic of epitopes. It may be shown that in fact one given epitope may be recognized by slightly different antibodies, and this contributes an extraordinary multiplication of the heterogeneity. This results from the clonal organization of lymphocytes, as originally proposed by Burnet (1). B lymphocytes (and T cells as well) are organized as discrete clones, each of which makes one and only one type of antibody molecule. In humans, the average population of lymphocytes at any given time is of the order of 1012 , of which approximately one-fifth are B cells and four-fifths are T cells. These B cells make about 1020 molecules of circulating immunoglobulins, a number that can be calculated from the Ig concentration in serum (10 mg/mL). The size of one clone is largely dependent on the immunization state and can vary from one single resting cell to several thousand or more. What happens when an animal is being immunized is that all clones that happen to interact somewhat with any potential epitope of the administered antigen will proliferate, expand, and synthesize as many different antibodies, thereby contributing to a very great heterogeneity. This of course made it very difficult, if not impossible, to study the fine structure of antibody molecules, especially when considering their amino acid sequences. A bias was brought to this by the existence of multiple myeloma in humans, or its equivalent as experimentally induced plasmacytoma in the mouse. These are lymphoproliferative disorders that affect plasma cells. As a result of the malignancy, one clone will expand specifically so that, very rapidly, it will become the major B-cell clone expressed in the body. The corresponding immunoglobulin will be secreted in excessively large amounts, and this provides a homogeneous material available for structural studies. An obvious drawback with this material is that the antibody specificity is generally unknown, which prevents its use for the study of antigen–antibody interactions. This difficulty was turned around when Kohler and Milstein (2) succeeded in making somatic hybrids between myeloma cells and normal B lymphocytes from hyperimmunized mice. These hybridomas combined the properties of myeloma cells, which can grow indefinitely in culture or upon transplantation in syngeneic strains of mice, with the clones of immunized lymphocytes that could be selected for a known antigenic specificity, leading to the fantastic expansion of “monoclonal antibodies” (mAbs). Monoclonal antibodies were not only suitable for detailed structural analysis of antibodies, including determination of the three-dimensional protein structure of antibody–antigen complexes. They also allowed access, through the hybridoma cells, to isolation of messenger RNA encoding Ig polypeptide chains and the corresponding complementary DNA, and thus to the complete gene organization of the three main Ig gene loci.
The basic structure of antibodies, elucidated from the pioneer work of Porter in England (3) and Edelman in the United States (4), is given by that of the IgG molecule—known initially as 7S-g-globulin. It is a symmetrical molecule, containing two identical heavy chains (H, 52 kDa and two identical light chains (L, 23 kDa), which are either kappa (k) or lambda (l) chains. Each H–L pair contains one combining site for the antigen, so the conventional H2 L2 IgG molecule is bivalent. The symmetry of the molecule is in agreement with the clonal selection theory, which postulated that each B cell expressed one and only one antibody specificity. This basic model could be extrapolated to other antibody classes, which differ from each other in the nature of their heavy chains and the degree of polymerization of the basic H2L2 unit. Five classes have thus been described in higher vertebrates: IgG, IgM, IgA, IgD, and IgE, having the corresponding g, m, a, d, and e heavy chains, respectively. IgM are expressed as H2L2 monomers at the cell surface of B cells, but are pentamers in the serum. IgA is mostly a dimer, whether other classes remain as monomers. The diversification of classes allows the antibodies to assume two types of function: (1) antigen recognition, common to all classes, and (2) biological or effector functions, such as complement fixation, active transplacental transfer, or fixation to various cell types, which amplify the action of antibodies and generally favor the elimination of a pathogen.
The functional duality of antibody molecules was clearly established in the late 1950 by Porter (5), who succeeded in cleaving the IgG antibody by papain, a proteinase, which led to the isolation of two identical fragments that were antigen binding, or Fab, and one Fc fragment that did not recognize the antigen but fixed the first component of the complement cascade. Structural support for this organization was clearly demonstrated by Hilschmann in 1965 (6), who reported the first amino acid sequence of two monoclonal human light chains. This is another example of the importance of monoclonal materials in this saga of the determination of the antibody structure. The work was performed on Bence-Jones proteins, which are free light chains isolated from the urine of patients with multiple myeloma. They are the result of a dysbalanced synthesis between the monoclonal heavy and light chains by the malignant plasma cells. Because of their smaller size, they pass the renal barrier and are thus easily isolated in large amounts from patient's urine. The major discovery was that the Bence-Jones light chains, or Ig light chains, were composed of one NH2-terminal half of 110 amino acid residues that profoundly differed between two patients, whereas the COOH-terminal half, also of 110 residues, was identical. This gave clear evidence for the existence of a huge structural diversity in antibodies, providing a unique basis for antibody specificity. Further extensive work indicated that heavy chains had also a variable region of similar size and a much longer constant region. The antibody combining site could thus be visualized as resulting from a contribution of both the VH and the VL regions. A more detailed analysis, computed by Kabat (7), indicated that within the V variable regions, subregions of hypervariability could be identified around positions 30, 50, and 100, which were later proven to participate directly in making up the antibody combining site. Three hypervariable regions, also called complementarity determining regions (CDR), were identified on each heavy and light chain, and their fine analysis revealed an extraordinary diversity that certainly could account for the expected huge repertoire of antibodies necessary to accommodate the potential repertoire of epitopes of the living world. Direct proof that the CDRs were implicated in antigen recognition came first from affinity labeling experiments, until a final confirmation was provide by X-ray crystallography analysis of crystals of antigen–antibody complexes. In fact, crystals were prepared from the Fab fragment of an antilysozyme antibody, combined with lysozyme, because the complete IgG antibody molecule contains a floppy “hinge” region that prevents crystallization. This structure, obtained by the group of Poljak (8), presented in Figure 1, indicates that about 20 amino acid residues of the antibody participate in binding the antigen, plus a similar number of residues from the antigen. On average, the area of interaction of the two partners is of the order of 600 Å2. Care should be taken in attempts to generalize this model to all other antigen-antibody pairs, because the size and shape of the antibody combining site varies greatly from one system to another. This variation is reflected in the wide variation of the association constants KA, between 105 and 1012 M–1 . The interactions between antigen and antibody are exclusively non covalent and involve primarily salt bridges, van der Waals interactions, and hydrogen bonds. The contributions of enthalpy and entropy vary immensely from one system to another, stressing again, if needed, the fantastic diversity potential of antibody molecules.
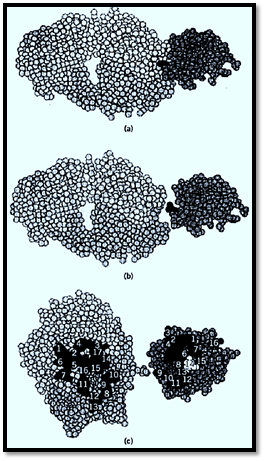
Figure 1. Three-dimensional structure of a lysozyme–Fab antilysozyme, showing the amino acid contributions from the heavy chain (upper part of Fab) and from the light chain (lower part). The antigen is on the right, and the Fab is on the left. (a, b) Side views. (c) Front view of the interacting residues of the antibody combining site (left) and epitope (right). (From Ref. 1, with permission from R. Poljak.)
Ultimately, the main problem that the immune system has to face is how to generate such a huge diversity with a number of genes that must necessarily be limited. At the time of the first data concerning the amino acid sequences of the light chains, many hypotheses were put forward to account for this diversity. There were two extremes: One considered that the diversity was exclusively the result of somatic hypermutations, whereas the other claimed that everything was encoded at the germline level, arguing that, provided that any light chain might pair with any heavy chain, 10,000 L-chain genes and 10,000 H-chain genes might generate 108 antibodies, a number that was already considered reasonable. Besides the fact that 20,000 genes would represent a large portion of the genome, this theory did not account for the conservation of the constant regions, a very serious objection that led Dreyer and Bennet (9) to propose that genes encoding the V and the C regions were separate in the germline. This was shown to be the case when the basic principles of the Ig gene organization were elucidated by the elegant experiments of Tonegawa in 1978 (10). In brief, V and C regions are encoded by separate regions within each of the three H, K, and L Ig loci, with a small number of C genes and a large number of V genes (Fig. 2). In addition, the V regions are in fact encoded by a mosaic of two gene segment clusters for the light chains (VL and JL) and three for the heavy chains (VH, D, and JH). Random combination of these elements takes place exclusively during B-cell differentiation and leads to a large collection of clones expressing various combinations from the basic gene mosaic. Diversity is further amplified greatly by other mechanisms, including somatic hypermutation. As a result, the number of distinct B-cell clones present at any time certainly far exceeds that necessary, especially in view of antigen–antibody recognition being partly degenerate.
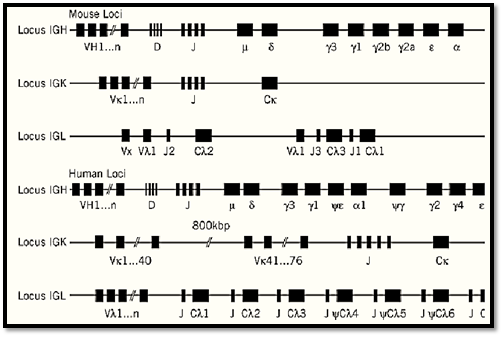
Figure 2. Schematic organization of the three Ig gene loci in (a mice and (b) humans. The IGK, IGL, and IGH loci, codi the k light chain, the l light chain, and the heavy chain, respectively, are located on mouse chromosomes 6, 16, and 12 an human chromosomes 2, 22, and 14, respectively.
References
1. M. F. Burnet (1959) The Clonal Selection Theory of Acquired Immunity. Vanderbilt University Press, Nashville, TN.
2. B. Köhler and C. Milstein (1975) Continuous culture of fused cells secreting antibody of predefined specificity. Nature 256, 495–499.
3. J. B. Fleischman, J. B. Pain, and R. R. Porter (1962) Reduction of gammaglobulins. Arch. Biochem. Biophys. Suppl. 1, 174–180.
4. G. M. Edelman and M. D. Poulik (1961) Studies on structural units of the -globulins. J. Exp. Med. 113, 861–884.
5. R. R. Porter (1959) The hydrolysis of rabbit gammaglobulin and antibodies by cristalline papain. Biochem. J. 73, 119–126.
6. N. Hilschmann and L. Craig (1965) Amino acid sequence studies with Bence-Jones proteins. Proc. Natl. Acad. Sci. 53, 1403–1409.
7. T. T. Wu and E. A. Kabat (1970) An analysis of the sequences of the variable regions of the Bence-Jones proteins and mycloma light chains and their implications for antibody complementarity. J. Exp. Med. 132, 211–250.
8. A. G. Amit, R. A. Mariuzza, S. E. V. Phillips, and R. J. Poljak (1986) Three-dimensional structure of an antigen–antibody complex at 2.8 A resolution. Science 233, 747–753.
9. W. J. Dreyer and J. C. Bennett (1965) The molecular basis of antibody formation: a paradox. Proc. Natl. Acad. Sci. USA 54, 864–869.
10. S. Tonegawa (1983) Somatic generation of antibody diversity. Nature, 302, 575–581.



|
|
|
|
إجراء أول اختبار لدواء "ثوري" يتصدى لعدة أنواع من السرطان
|
|
|
|
|
|
|
دراسة تكشف "سببا غريبا" يعيق نمو الطيور
|
|
|
|
|
|
لأعضاء مدوّنة الكفيل السيد الصافي يؤكّد على تفعيل القصة والرواية المجسّدة للمبادئ الإسلامية والموجدة لحلول المشاكل المجتمعية
|
|
|
|
قسم الشؤون الفكرية يناقش سبل تعزيز التعاون المشترك مع المؤسّسات الأكاديمية في نيجيريا
|
|
|
|
ضمن برنامج عُرفاء المنصّة قسم التطوير يقيم ورشة في (فنّ الٕالقاء) لمنتسبي العتبة العباسية
|
|
|
|
وفد نيجيري يُشيد بمشروع المجمع العلمي لحفظ القرآن الكريم
|