

 النبات
النبات
 الحيوان
الحيوان
 الأحياء المجهرية
الأحياء المجهرية
 علم الأمراض
علم الأمراض
 التقانة الإحيائية
التقانة الإحيائية
 التقنية الحيوية المكروبية
التقنية الحيوية المكروبية
 التقنية الحياتية النانوية
التقنية الحياتية النانوية
 علم الأجنة
علم الأجنة
 الأحياء الجزيئي
الأحياء الجزيئي
 علم وظائف الأعضاء
علم وظائف الأعضاء
 الغدد
الغدد
 المضادات الحيوية
المضادات الحيوية|
Read More
Date: 14-12-2015
Date: 4-6-2021
Date: 7-12-2015
|
Replication
There is no more critical issue in the origin of life than a method for the faithful and timely replication of genetic information. Genes are encoded in deoxyribonucleic acid (DNA), which is made of four types of nucleotides, distinguished by the bases adenine (A), guanine (G), cytosine (C), and thymine (T). When James Watson and Francis Crick discovered the double helical structure of deoxyribonucleic acid (DNA), they also recognized that DNA replication could occur by opening the double helix into single strands, and from those templates creating new complementary strands by the principle of base pairing. As an overview, their proposal was correct. The details of the chemistry, of the enzymes involved, and of the structure on which replication occurs tell a fascinating story that is far more complex than Watson and Crick anticipated.
Semiconservative Replication
Chromosomes are the extended molecules of DNA that carry genes in both bacteria and eukaryotes. Bacterial chromosomes are usually circular, with the double helix looping around to make a complete circle. Eukaryotic chromosomes are linear, with the double helix sealing up at the two distant ends. In both cases, the result of replication is that one double helix with its two complementary strands of nucleotides becomes two identical double helices with the same sequence of nucleotides. In this way, the genetic material of a cell is passed along unchanged through all the descendants of the original cell (except for replication errors or other mutations).
Each new helix includes one original side and one new side, and so the replication process is said to be “semiconservative.” Semiconservative replication was discovered by Matthew Meselson and Frank Stahl, who grew bacteria with radioactive nucleotides. Fully radioactive chromosomes became half as radioactive after one round of replication, indicating that half the original chromosome was preserved in each new copy.
Replication Forks and Accessory Enzymes
Replication is a huge task, whether in bacteria or in eukaryotes. There are several physical and biochemical challenges the cell must overcome. First, the site or sites at which to begin replication must be located and the proper enzymes collected there. Second, the double helix must be unwound to expose the two strands. This imposes twisting strain on the portions of the helix farther away from the unwinding site, much like untangling a twisted phone cord does, and those forces must be relieved to prevent breakage of the DNA strands. Complementary nucleotides must be put in place and linked to form a new strand, and errors must be checked and corrected.
The orientation of the two strands poses an additional challenge. Because of the way the deoxyribose sugar is structured, the sugar, and hence the whole DNA strand, has a direction, an up versus down, so to speak. The two sides are oriented with up and down directions opposite, in a so-called antiparallel fashion. In biochemical terms, one direction is 5'-3' (“five-prime to three-prime”), while the other side is oriented 3'-5'. The consequence of this arises because the enzymes that perform replication only function in one direction. The solution to this problem is discussed below.
Much of the understanding of DNA replication has come from studying the bacterium Escherichia coli. In bacterial chromosomes one site, called the origin of replication, has a sequence of base pairs to which an initiator protein binds. The initiator protein attracts enzymes called helicases that interrupt base pairing in a way that separates the double helix into a short region of two single strands. Other binding proteins then attach to maintain the single strand separation on the double-stranded region. Adjacent to the single-stranded region, an enzyme called gyrase cleaves and reforms the sugar-phosphate backbones in the double strand helix, which relieves the strain that the single strand separation causes. Separating the double stranded helix into a region of two single strands creates a “replication bubble” initially of a few hundred nucleotides. Within the replication bubble, each strand serves as the template for a new chain; these chains grow in opposite directions because the templates have opposite 5'to 3'polarity. As the chains elongate, the replication bubble expands in both directions, and the ends become known as replication forks. As replication occurs, the helicase molecules are pushed toward the fork where the double strand separates into single strands, thus moving the fork and extending the region of single strands.

Figure 1. Replication adds a free nucleotide to the 3' OH group of the last nucleotide in the growing chain of DNA.
Priming and Elongation
E. coli use three enzymes for replications, called pol I, pol II, and pol III. Early work with pol I showed that replication adds a free nucleotide to the 3'OH group of the last nucleotide in the growing chain (see Figure 1). Each nucleotide-added hydrogen bond to the complementary nucleotide in the template single strand. The 3'OH of the last nucleotide attacks the high- energy triphosphate group at the 5'position of the free nucleotide, splitting off two phosphates and forming a covalent bond to the innermost phosphate. This binds the new nucleotide to the existing chain.
Some anticancer and antiviral drugs are nucleotides missing the 3' OH. Such “dideoxy” nucleotides shut down replication after being incorporated into the strand. Fast-replicating DNA in cancer cells or viruses is inactivated by these drugs.
To begin elongation, DNA polymerases require an existing chain with a 3' OH end. This posed the problem of how replication could ever begin, since the needed 3' OH is on the newly replicated strand. The key to understanding the initiation of replication came with the discovery of ribonucleic acid (RNA) priming, which does not require an existing 3' OH to start the process. In priming, an enzyme called primase places a short sequence of RNA nucleotides into position at the origin of replication. This sequence is complementary to the 3'end of the single-stranded portion of the template at that point. DNA polymerase then adds nucleotides to the RNA’s 3'OH, continuing replication, until they reach the end of the complementary template.
Since polymerase only elongates in a 5'to 3'direction, at each replication fork only one chain is elongating in a smooth, unbroken fashion. The

Figure 2. The proteins at a DNA replication fork.
continuous elongation product is termed the “leading strand,” while the discontinuous product is the “lagging strand.” For every new section on the lagging strand, primase creates a new RNA primer and 3'OH end to start DNA chain elongation. Called Okazaki fragments after their discoverer, each short fragment lasts only a brief period before the primer ribonucleotides are digested and replaced with DNA nucleotides, which are then ligated (“tied”) to the adjacent nucleotides by DNA ligase. Ligation of the Okazaki fragments into long chains completes synthesis of the lagging strand (see Figure 2). The pol III enzyme is a large complex of subunits that catalyzes both leading-strand and lagging-strand elongation and has roles in proofreading and replacing mismatched nucleotides. The pol I enzyme digests the RNA nucleotides and replaces them with DNA nucleotides; it also proofreads and replaces incorrect with correct nucleotides. It is thought that pol II mostly repairs replication errors.
Replication in the E. coli chromosome is bidirectional and continues in opposite directions until the two replication forks meet about halfway around the circular chromosome. Replication is then complete.
Special Features of Eukaryotic Replication
Replication of eukaryotic chromosomes is more complex inasmuch as they are linear (versus circular) and usually much larger than bacterial chromosomes. DNA replication is restricted to the S, or synthesis, phase during the cell cycle, between mitotic divisions. As a result of replication, each chromosome consists of two identical chromatids joined together, which are then separated into daughter cells by mitotic division.
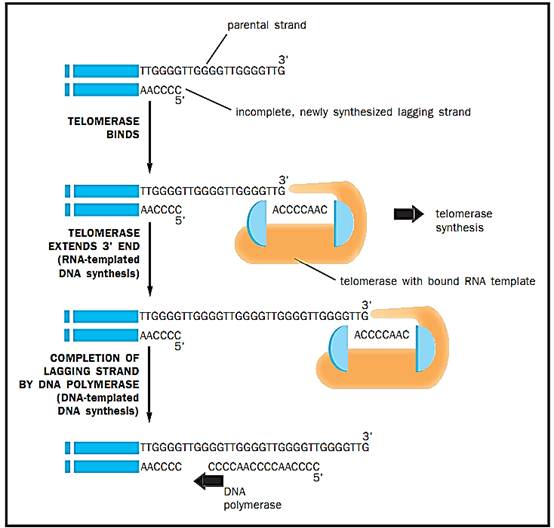
Figure 3. Telomerase is a hybrid protein-RNA molecule; the RNA sequence is complementary to several repeat lengths of the telomeric DNA. The telomerase uses the RNA sequence to bind to the template end of telomeric DNA, and uses the overhang protein portion to add DNA nucleotides to the template, extending it beyond its normal length.
Instead of a single origin of replication, as in bacteria, eukaryotic chromosomes have many origins for each chromosome in keeping with their much larger size. Replication proceeds bidirectional from each origin until it meets the replication fork from the adjacent origin. A replicon refers to the interval replicated from one origin. This concept is shaky, however, since there is evidence that origins of replication are somewhat specific to the stage and tissue the cells are in, rather than being a permanent physical property. For example, nuclei in the Drosophila embryo divide every nine minutes initially. This requires the fastest replication of chromosomes known and utilizes many origins. Later stages use fewer origins.
Eukaryotes have five polymerases, termed alpha, beta, gamma, delta, and epsilon. Replication of nuclear DNA utilizes the alpha and delta polymerases. Alpha polymerase is a complex of several subunits, one of which has primase activity when it is in the complex. The alpha polymerase is thought to carry out synthesis of the lagging strand, whereas the delta polymerase, also a complex of subunits but lacking primase activity, carries out synthesis of the leading strand. As in the prokaryotes, helicase and gyrase are required to unwind the double helix ahead of the replication fork. The alpha and delta polymerases function in proofreading and correction as well. The beta and epsilon polymerases are thought to carry out nuclear DNA repair. The gamma polymerase replicates the mitochondrial genome. It lacks the error correction mechanism of the other polymerases, with the re sult that the mutation rate in mitochondrial replication is substantially higher than it is in replication of nuclear DNA.
Telomeres and Telomerase
Because eukaryotic chromosomes are linear and have ends, another enzyme, called telomerase, is necessary but is not found in the prokaryotes. The problem with chromosome ends, called telomeres, is that the 3'template for the lagging strand cannot be primed at the last nucleotides because there is no further DNA on which to build. By itself, this would reduce the length of the lagging strand and the chromosome would get shorter at each replication. Telomeric DNAs, which are repeats of a specific sequence of six nucleotides, are normally present at the ends of chromosomal DNA and avoid this problem. Telomerase is a hybrid protein-RNA molecule; the RNA sequence is complementary to several repeat lengths of the telomeric DNA. The telomerase uses the RNA sequence to bind to the template end of telomeric DNA and uses the overhang protein portion to add DNA nucleotides to the template, extending it beyond its normal length. With several movements of the enzyme outward and reiterations of this process, the template 3'end is extended sufficiently to allow DNA polymerase to complete synthesis of a normal length lagging strand. In multicellular organisms, somatic cells usually cease mitotic division during development and lack telomerase activity thereafter. Cancer cells abnormally turn their telomerase back on, which enables the cell to divide continually. Telomerase is a target of drug research for the combat of cancer.
References
Cotterill, Sue, ed. Eukaryotic DNA Replication: A Practical Approach. Oxford: Oxford University Press, 1999.
Kornberg, Arthur, and Tania Baker. DNA Replication. New York: W. H. Freeman and Company, 1991



|
|
|
|
"عادة ليلية" قد تكون المفتاح للوقاية من الخرف
|
|
|
|
|
|
|
ممتص الصدمات: طريقة عمله وأهميته وأبرز علامات تلفه
|
|
|
|
|
|
|
المجمع العلمي للقرآن الكريم يقيم جلسة حوارية لطلبة جامعة الكوفة
|
|
|