

النبات

مواضيع عامة في علم النبات

الجذور - السيقان - الأوراق

النباتات الوعائية واللاوعائية

البذور (مغطاة البذور - عاريات البذور)

الطحالب

النباتات الطبية


الحيوان

مواضيع عامة في علم الحيوان

علم التشريح

التنوع الإحيائي

البايلوجيا الخلوية


الأحياء المجهرية

البكتيريا

الفطريات

الطفيليات

الفايروسات


علم الأمراض

الاورام

الامراض الوراثية

الامراض المناعية

الامراض المدارية

اضطرابات الدورة الدموية

مواضيع عامة في علم الامراض

الحشرات


التقانة الإحيائية

مواضيع عامة في التقانة الإحيائية


التقنية الحيوية المكروبية

التقنية الحيوية والميكروبات

الفعاليات الحيوية

وراثة الاحياء المجهرية

تصنيف الاحياء المجهرية

الاحياء المجهرية في الطبيعة

أيض الاجهاد

التقنية الحيوية والبيئة

التقنية الحيوية والطب

التقنية الحيوية والزراعة

التقنية الحيوية والصناعة

التقنية الحيوية والطاقة

البحار والطحالب الصغيرة

عزل البروتين

هندسة الجينات


التقنية الحياتية النانوية

مفاهيم التقنية الحيوية النانوية

التراكيب النانوية والمجاهر المستخدمة في رؤيتها

تصنيع وتخليق المواد النانوية

تطبيقات التقنية النانوية والحيوية النانوية

الرقائق والمتحسسات الحيوية

المصفوفات المجهرية وحاسوب الدنا

اللقاحات

البيئة والتلوث


علم الأجنة

اعضاء التكاثر وتشكل الاعراس

الاخصاب

التشطر

العصيبة وتشكل الجسيدات

تشكل اللواحق الجنينية

تكون المعيدة وظهور الطبقات الجنينية

مقدمة لعلم الاجنة


الأحياء الجزيئي

مواضيع عامة في الاحياء الجزيئي


علم وظائف الأعضاء


الغدد

مواضيع عامة في الغدد

الغدد الصم و هرموناتها

الجسم تحت السريري

الغدة النخامية

الغدة الكظرية

الغدة التناسلية

الغدة الدرقية والجار الدرقية

الغدة البنكرياسية

الغدة الصنوبرية

مواضيع عامة في علم وظائف الاعضاء

الخلية الحيوانية

الجهاز العصبي

أعضاء الحس

الجهاز العضلي

السوائل الجسمية

الجهاز الدوري والليمف

الجهاز التنفسي

الجهاز الهضمي

الجهاز البولي


المضادات الميكروبية

مواضيع عامة في المضادات الميكروبية

مضادات البكتيريا

مضادات الفطريات

مضادات الطفيليات

مضادات الفايروسات

علم الخلية

الوراثة

الأحياء العامة

المناعة

التحليلات المرضية

الكيمياء الحيوية

مواضيع متنوعة أخرى

الانزيمات
The Path of Nucleosomes in the Chromatin Fiber
المؤلف:
JOCELYN E. KREBS, ELLIOTT S. GOLDSTEIN and STEPHEN T. KILPATRICK
المصدر:
LEWIN’S GENES XII
الجزء والصفحة:
25-3-2021
2953
The Path of Nucleosomes in the Chromatin Fiber
KEY CONCEPTS
- The primary structure of chromatin is a 10-nm fiber that consists of a string of nucleosomes.
- The secondary structure of chromatin is formed by interactions between neighboring nucleosomes that promote formation of more condensed fibers.
- 30-nm fibers are a prevalent type of secondary structure that contain 6 nucleosomes/turn, organized into either a one-start solenoid or a two-start zigzag helix.
- Histone H1, histone tails, and increased ionic strength all promote the formation of secondary structures, including the 30-nm fiber.
- Secondary chromatin fibers are folded into higher-order, three-dimensional structures that comprise interphase or mitotic chromosomes.
When chromatin is released from nuclei and examined with an electron microscope, we can see two types of fibers: the 10-nm fiber and the 30-nm fiber. They are described by the approximate diameter of the thread (that of the 30-nm fiber actually varies from around 25–30 nm). The 10-nm fiber is essentially a continuous string of nucleosomes and represents the least compacted level of chromatin structure. In fact, a stretched-out 10-nm fiber resembles a string of beads in which we can clearly distinguish nucleosomes connected by linker DNA, as demonstrated in FIGURE 1. The 10-nm fiber structure is obtained under conditions of low ionic strength and does not require the presence of histone H1. This means that it is a function strictly of the nucleosomes themselves. FIGURE 2 shows a depiction of the continuous series of nucleosomes in this fiber.
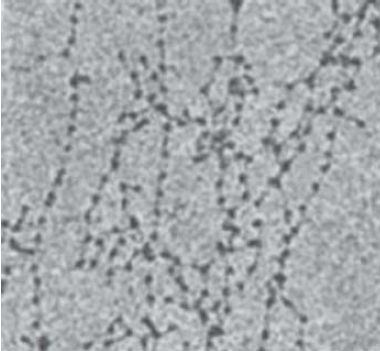
FIGURE 1 The 10-nm fiber in partially unwound state can be seen to consist of a string of nucleosomes.
Photo courtesy of Barbara Hamkalo, University of California, Irvine.
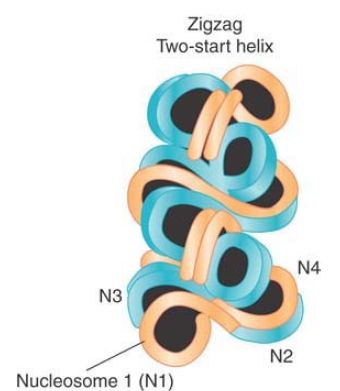
FIGURE 2 The 30-nm fiber is a two-start helix consisting of two rows of nucleosomes coiled into a solenoid.
Reprinted from Cell, vol. 128, D. J. Tremethick, Higher-order structure of chromatin …, pp. 651–654. Copyright 2007, with permission from Elsevier [http://www.sciencedirect.com/science/journal/00928674].
When chromatin is visualized in conditions of greater ionic strength, the 30-nm fiber is obtained. An example is given in FIGURE 3. You can see that the fiber has an underlying coiled structure. It has approximately 6 nucleosomes for every turn, which corresponds to a packing ratio of 40 (i.e., each mm along the axis of the fiber contains 40 mm of DNA). The formation of this fiber requires the histone tails, which are involved in internucleosomal contacts, and is facilitated by the presence of a linker histone such as H1.
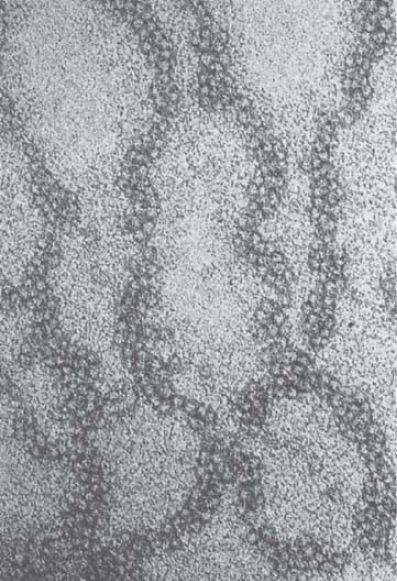
FIGURE 3 The 30-nm fiber has a coiled structure.
Photo courtesy of Barbara Hamkalo, University of California, Irvine.
Nucleosomes are arranged into a helical array within the 30-nm fiber, with the linker DNA occupying the central cavity. The two main forms of this helical structure are a single start solenoid, which forms a linear array, and a two-start zigzag that in effect consists of a double row of nucleosomes. FIGURE 4 shows a two-start model suggested by crosslinking data identifying a double stack of nucleosomes in the 30-nm fiber. Although this model is also supported by the crystal structure of a tetranucleosome complex, recent studies suggest that the type of helical structure (e.g., one-start solenoid or two-start zigzag) is influenced by the length of linker DNA within the 10-nm fiber. Furthermore, biochemical studies suggest that 30-nm fibers might contain a heterogeneous mixture of one-start and two-start helical organizations, rather than a single, uniform structure.
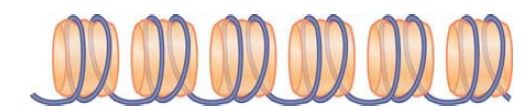
FIGURE 4 The 10-nm fiber is a continuous string of nucleosomes.
Levels of folding beyond the 30-nm fiber are very poorly understood, but it has long been believed that the 40-fold compaction provided by the 30-nm fiber is still a long way from the levels of compaction required for interphase or mitotic packaging of chromosomes. Researchers have observed chromatin fibers with diameters of 60–300 nm (called chromonema fibers) by both light and electron microscopy. Such fibers were presumed to consist of folded 30-nm fibers and would represent a major level of compaction (a 30-nm fiber running just across the width of a 100- nm fiber would contain more than 10 kb of DNA), but the actual substructures of these large fibers remain unknown. Indeed, recent microscopy studies do not detect significant levels of 30-nm fibers within chromatin in situ, suggesting that 30-nm fibers might exist only in regions of low chromatin density (or maybe not at all!). In contrast, several studies have provided compelling evidence that even highly condensed mitotic chromatin might be composed of only 10-nm fibers, densely packed into an interdigitated “polymer melt” or “fractal globule.” This type of organization facilitates a dense packaging of DNA while preserving the ability to fold and unfold genomic loci. FIGURE 5 shows a hypothetical depiction of this higher-order folding model.
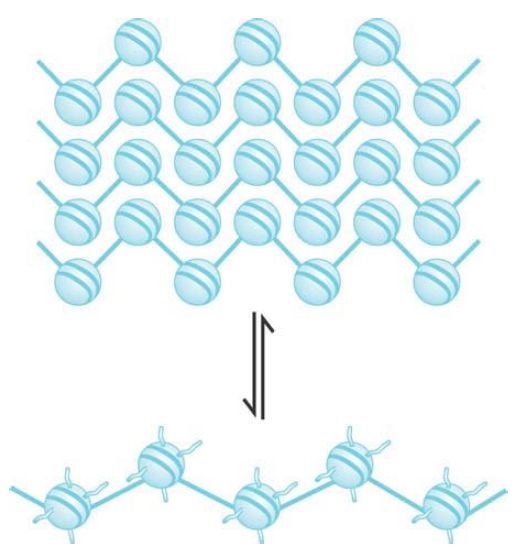
FIGURE 5 A model for higher order chromatin structure involving interdigitation of 10-nm chromatin fibers. The resulting fractal globule allows for reversible extrusion of individual fibers for nuclear functions such as transcription.
How can genomic DNA fit into the nuclear volume if organization into 10-nm fibers provides only a 6-fold compaction ratio?
Historically, we have thought about DNA packaging into the nucleus from the point of view of linear compaction—if DNA is stretched end-to-end, it must be shortened by about 10,000-fold to form a mitotic chromosome. This led to the popular idea of hierarchical levels of chromatin folding (e.g., 10-nm → 30-nm → 60- to 300-nm fibers). However, if genomic DNA is modeled as a simple cylinder, the volume of DNA in a diploid mammalian nucleus is actually less than 6% of the nuclear volume. Wrapping DNA around histones actually takes up more space! In this view, the role of chromatin organization is not to compact linear DNA into the nuclear space, rather it is to help oppose the negative charge of DNA and facilitatethe folding and bending of DNA on itself. In this view, the extended 10-nm fiber is highly flexible and can not only bend and kink but also self-associate to form dense networks that satisfy nuclear packaging requirements.
 الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
 اخر الاخبار
اخر الاخبار
اخبار العتبة العباسية المقدسة

الآخبار الصحية















 قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة
قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة "المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة
"المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة (نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)
(نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)


















