

النبات

مواضيع عامة في علم النبات

الجذور - السيقان - الأوراق

النباتات الوعائية واللاوعائية

البذور (مغطاة البذور - عاريات البذور)

الطحالب

النباتات الطبية


الحيوان

مواضيع عامة في علم الحيوان

علم التشريح

التنوع الإحيائي

البايلوجيا الخلوية


الأحياء المجهرية

البكتيريا

الفطريات

الطفيليات

الفايروسات


علم الأمراض

الاورام

الامراض الوراثية

الامراض المناعية

الامراض المدارية

اضطرابات الدورة الدموية

مواضيع عامة في علم الامراض

الحشرات


التقانة الإحيائية

مواضيع عامة في التقانة الإحيائية


التقنية الحيوية المكروبية

التقنية الحيوية والميكروبات

الفعاليات الحيوية

وراثة الاحياء المجهرية

تصنيف الاحياء المجهرية

الاحياء المجهرية في الطبيعة

أيض الاجهاد

التقنية الحيوية والبيئة

التقنية الحيوية والطب

التقنية الحيوية والزراعة

التقنية الحيوية والصناعة

التقنية الحيوية والطاقة

البحار والطحالب الصغيرة

عزل البروتين

هندسة الجينات


التقنية الحياتية النانوية

مفاهيم التقنية الحيوية النانوية

التراكيب النانوية والمجاهر المستخدمة في رؤيتها

تصنيع وتخليق المواد النانوية

تطبيقات التقنية النانوية والحيوية النانوية

الرقائق والمتحسسات الحيوية

المصفوفات المجهرية وحاسوب الدنا

اللقاحات

البيئة والتلوث


علم الأجنة

اعضاء التكاثر وتشكل الاعراس

الاخصاب

التشطر

العصيبة وتشكل الجسيدات

تشكل اللواحق الجنينية

تكون المعيدة وظهور الطبقات الجنينية

مقدمة لعلم الاجنة


الأحياء الجزيئي

مواضيع عامة في الاحياء الجزيئي


علم وظائف الأعضاء


الغدد

مواضيع عامة في الغدد

الغدد الصم و هرموناتها

الجسم تحت السريري

الغدة النخامية

الغدة الكظرية

الغدة التناسلية

الغدة الدرقية والجار الدرقية

الغدة البنكرياسية

الغدة الصنوبرية

مواضيع عامة في علم وظائف الاعضاء

الخلية الحيوانية

الجهاز العصبي

أعضاء الحس

الجهاز العضلي

السوائل الجسمية

الجهاز الدوري والليمف

الجهاز التنفسي

الجهاز الهضمي

الجهاز البولي


المضادات الميكروبية

مواضيع عامة في المضادات الميكروبية

مضادات البكتيريا

مضادات الفطريات

مضادات الطفيليات

مضادات الفايروسات

علم الخلية

الوراثة

الأحياء العامة

المناعة

التحليلات المرضية

الكيمياء الحيوية

مواضيع متنوعة أخرى

الانزيمات
DNA Ligase
المؤلف:
M. J. Engler and C. C. Richardson
المصدر:
In The Enzymes
الجزء والصفحة:
25-4-2016
4415
DNA Ligase
1. Physiological Functions
Several fundamental biological processes involve one or more steps in which an enzyme, DNA ligase, catalyzes the covalent joining of one piece of DNA to another (1-3). For example, DNA altered spontaneously or by environmental causes is restored to its original base sequence by DNA repair. DNA is duplicated by DNA replication and rearranged by homologous recombination. All these processes require DNA ligase activity to link preformed pieces of DNA to one another.
During one form of DNA repair, damaged nucleotides are removed by enzymes to leave ultimately a gap in one strand of the duplex DNA, which is then filled by a DNA polymerase to form a product with a discontinuity (nick) between two adjacent nucleotides in the phosphodiester backbone. Likewise, semidiscontinuous DNA replication leads to the formation of Okazaki Fragments that abut one another on the complementary parental DNA template strand so that a nick exists between their termini. Finally, pieces of DNA from two different chromosomes become associated during recombination so that similar nicked duplexes are formed in each chromosome. DNA ligase catalyzes the covalent joining of the DNA strands to seal the nicks and form intact, recombined duplexes. The enzyme is involved in other functions as well; whenever two pieces of DNA must be covalently joined or a DNA molecule circularized, DNA ligase probably catalyzes the reaction.
DNA fragments with appropriate termini that are aligned on a complementary strand so that their ends are juxtaposed can be joined by DNA ligase to form a phosphodiester bond (Fig. 1). For effective joining, the two strands (A and B) must be directly abutted without a gap (no missing base pairs) and must also have a 3′-OH on strand A and a 5′-PO4 on strand B. The 3′-hydroxyl (-OH) of fragment A is ligated to the 5′-PO 4 of B to form a 3′ → 5′ phosphodiester bond. These two strands are abutted by base pairing with a complementary, intact strand (C). Since the formation of a phosphodiester bond is endergonic, ATP or NAD+, depending on the source of the enzyme, acts as a cofactor to supply the energy.
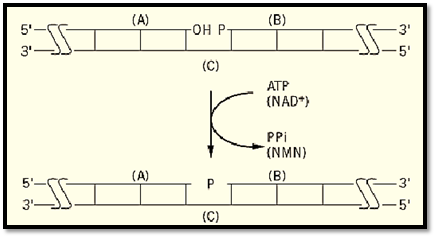
Figure 1. The overall DNA ligase reaction. A nick with a 3′-hydroxyl (- OH) on strand (A) adjacent to a 5′-phosphate (P- ) terminated strand (B) on a complementary strand (C) is converted to an intact phosphodiester bond- )P- ). ATP or NAD+ supply the energy for bond formation, and PPi or NMN, respectively, is the other product of the reaction. Long horizontal lines represent DNA strands with the polarity indicated; vertical lines represent base pairs.
2. Distribution
All free-living organisms are believed to produce one or more DNA ligases. Until recently, bacteria were thought to contain a single DNA ligase that performed all their DNA joining reactions. Yeast appears to have two DNA ligases. Some bacteriophage and animal viruses also encode DNA ligases. Higher eukaryotes probably contain four different DNA ligases that are specialized to perform particular tasks. The DNA ligases from animal and plant cells, archaebacteria, bacteriophage, and animal viruses use ATP as the energy-supplying cofactor for the ligation reaction. Eubacteria, with a single reported exception, use NAD+ for this purpose. Cheng and Shuman (4) have shown that Haemophilus influenzae contains an ATP-dependent ligase, in addition to the expected NAD+-dependent enzyme. Whether this finding represents an anomaly in the general cofactor-utilization classification scheme awaits the demonstration of additional ATP-dependent activities in other eubacteria. Searches of the sequences of the genomes of several other eubacteria reveal potential open reading frames that share homologies with both the expected NAD+-dependent enzymes and with ATP-dependent DNA ligases. It will be important to determine whether any of these organisms also produce ATP-dependent DNA ligases and, if so, how they relate functionally to the NAD+-dependent enzymes.
DNA ligase is essential because of its role in the crucial DNA transactions of the cell. Cells containing conditional mutants of the enzyme in Escherichia coli are lethal when the ligase activity is severely inactivated. They display abnormal replication, repair, and recombination phenotypes. Because E. coli cells normally contain many copies of the enzyme, small amounts of residual activity can maintain viability. The enzymes in bacteria seem to act independently in their multiple roles, that is, not in specific association with other proteins. For example, the NAD+-dependent enzyme of E. coli can substitute for the inactivated bacteriophage ATP-dependent DNA ligase of an infecting bacteriophage. In eukaryotes, specific proteins may associate with DNA ligases to form complexes involved in particular functions.
3. Properties
The DNA ligases comprise single polypeptide chains with molecular weights ranging from approximately 40 kDa to greater than 100 kDa, and they probably act as monomers. The genes of many DNA ligases have been sequenced, and several have been overexpressed and the enzymes isolated, purified, and characterized in detail. Several bacterial and bacteriophage DNA ligases are commercially available. Analyses of the sequences of genes encoding DNA ligases reveals conserved amino-acid sequence motifs among the ATP-dependent enzymes and other conserved regions among the NAD+-dependent group. Sequence comparisons also reveal that the ATP-dependent DNA ligases are part of a superfamily that includes RNA ligases and RNA capping enzymes (5). The structure of only the bacteriophage T7 DNA ligase, a small, ATP-dependent enzyme, has been determined (6). It forms a bilobal structure that is reminiscent of the structures of the methyltransferases used for DNA modification.
4. Mechanism
Phosphodiester bond formation between template-aligned, abutted 3′-OH and 5-PO4 termini proceeds in three steps, with the formation of two covalent intermediates (Fig. 2). One of these intermediates involves the enzyme, the other the DNA. In the first step of the reaction, which can occur in the absence of DNA, the enzyme reacts with ATP (or NAD+) to form a covalent enzyme–AMP complex. The adenylyl group is joined to the -amino group of a lysine residue at the active site by a phosphorus–nitrogen (phosphoamide) linkage that has a large free energy of hydrolysis (Fig. 2a). If ATP is the cofactor, PPi is released; if NAD+ serves as the cofactor, nicotinamide mononucleotide (NMN) is released in this step. The free energy of hydrolysis of a phosphoanhyride bond in the ATP or NAD+ is used to form the enzyme-bound, activated adenylyl group.
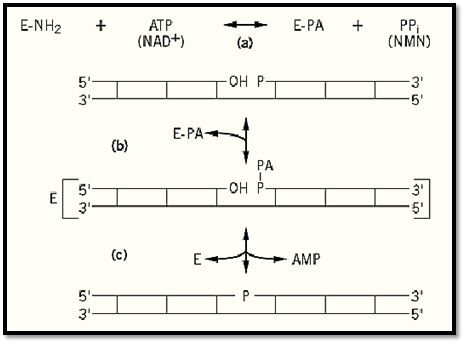
Figure 2. The DNA ligase reaction mechanism. (a) The enzyme [E–NH2] is adenylylated on a specific lysine residue [E–PA] by reaction with ATP (or NAD+) with the release of PPi (or NMN). (b) The DNA is adenylylated [AP- P -] by transfer of the adenylyl group from the enzyme to the 5′-PO4 at the nick. (c) The enzyme catalyzes an attack of the 3′-OH at the nick on the activated phosphate, to form the 3′ → 5′ phosphodiester bond and release the enzyme. Long horizontal lines represent DNA strands with the polarity indicated; vertical lines represent base pairs.
In the next step, the second covalent intermediate is formed when the adenylyl group is transferred from the enzyme to the terminal 5′-PO 4 at the DNA nick, to form an AMP (5′ → 5′) DNA phosphoanhydride linkage (Fig. 2b). In the final step of the reaction, which requires the free )unadenylylated) form of the enzyme, the AMP is displaced from the DNA by a nucleophilic attack of the 3′-OH of the adjacent DNA fragment to form the phosphodiester bond and to release the AMP from the DNA Fig. 2c).
Both covalent intermediates in the reaction have been isolated and are kinetically competent in the reaction. The reaction requires a divalent metal cation, most likely Mg2+ in the cell. Steady-state kinetic analysis of the E. coli enzyme shows that it follows ping-pong kinetics—consistent with the described mechanism. The overall reaction is reversible. The first step is readily reversed, as indicated by the facile ATP–PPi or NAD+ NMN isotope exchange reactions catalyzed. The enzyme also catalyzes an AMP-dependent nicking–closing (topoisomerase( activity that will relax positively or negatively supercoiled DNA, indicating that the second step is also reversible.
The nature of the ends of DNA fragments affects the ability of DNA ligase to join them together. For example, duplex substrates with so called cohesive, “sticky” ends are readily joined by all DNA ligases (Fig. 3a). These substrates are formed from DNA fragments that contain short, complementary single-stranded extensions at their termini; two or more such nucleotides are required for efficient joining. These extensions can be of either strand, so long as they allow the two duplexes to anneal to one another and provide the requisite abutted 3′-OH and 5′-PO4 termini. The ends of these duplex fragments hybridize to form a substrate in which there is a ligatable discontinuity in each strand. In essence, the complementary strands serve to convert the intermolecular joining of the two DNA pieces into an intramolecular reaction by bringing them together. Another class of duplex substrates, specifically, those with blunt ends (Fig. 3b), which have no single-stranded extensions, are much less efficiently ligated, because the fragments to be joined are not brought into proximity by a template strand. The efficiency of this intermolecular blunt-end joining reaction depends greatly on the particular ligase. Bacteriophage T4 ligase performs the reaction best and has been extensively used for this purpose, whereas the enzyme from E. coli will join blunt-end duplexes only in the presence of excluded-volume agents, such as polyethylene glycol. Other DNA structures, such as those with noncomplementary, protruding ends or nonabutted fragments creating a gap on a template strand, can be ligated, but inefficiently. T4 DNA ligase is again more tolerant of these unusual structures than is the bacterial enzyme.
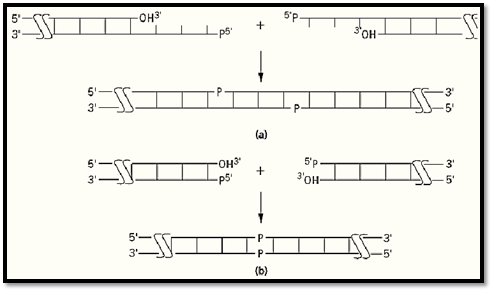
Figure 3. Common DNA ends joined by DNA ligase. (a) Fragments with a 5′ extension (shown here) or with a 3′ extens and complementary bases (sticky ends) form a ligatable structure. (b) Ligation of blunt ends. Long horizontal lines repre DNA strands with the polarity indicated; vertical lines represent base pairs.
5. Polynucleotide Substrates
The physiological polynucleotide substrates for DNA ligase are predominantly DNA chains brought into proximity by a third, complementary DNA template strand as shown in Figure 1. Studies of various combinations of RNA and DNA substrates reveal that DNA ligases will also sometimes use RNA in vitro. RNA can be joined to RNA on either a DNA or RNA template. RNA with a 3′-OH can also be joined to DNA with a 5′-PO4 on a DNA template. Likewise, DNA with a 3′-OH can be joined to RNA with a 5′-PO4 on a DNA template. The exact combinations of oligonucleotide types and the efficiencies of the joinings again depend on the particular enzyme used and the reaction conditions employed. T4 DNA ligase is again often more permissive in these “unnatural” ligations than is the E. coli enzyme. On binding to the DNA, the ATP-dependent enzymes may specifically recognize a nick in the duplex that contains the requisite 3′-OH and 5′-PO4 termini (7). Vaccinia virus DNA ligase appears to require a B-DNA helix to the 5′ side of the nick, suggesting that substrate conformation is another parameter that can influence recognition (8). It should be noted that some studies of substrate requirements were carried out with homopolymeric substrates and that the results might be quantitatively different for substrates containing heterogeneous sequences.
6. Assays
Several assays have been used to quantify DNA ligase activity. [ 3H]poly d(A-T) folds on itself to form base pairs that create a dumbbell-like structure with a nick that can be sealed by DNA ligase to form a covalently closed, single-stranded circle. This product of the ligase reaction is resistant to exonuclease III, whereas the unsealed substrate is hydrolyzed to deoxyribonucleotides by the exonuclease. The rejoining, as determined by gel electrophoresis, of a mixture of fragments formed by a restriction enzyme digestion of a plasmid is a common, qualitative assay. Synthetic oligodeoxyribonucleotides allow the scheme outlined in Figure 1 to serve as an assay. If strand A is labeled at its 5′ end with 32P, DNA ligase will convert the radiolabeled 5′-phosphate into a product with a phosphomonoester-resistant bond that can be quantified directly; alternatively, the longer product can be detected by gel electrophoresis. Analogously, [5′-32 P]d(pT)n oligonucleotides aligned on a long poly d(A) template serve as substrates for a similar assay in which the products can be quantified. The restoration of transforming activity to DNA that has been partially inactivated by deoxyribonuclease treatment has also served as an assay. Finally, the adenylylated enzyme formed in the first step of the reaction can be assayed by using an appropriately radiolabeled ATP or NAD+ and determining the acid-precipitable, protein-bound label, or by observing the difference in migration of the free and adenylylated enzyme during polyacrylamide electrophoresis. The first step of the reaction can also be assayed by measuring an ATP–PPi or NAD+-NMN exchange reaction.
7. Applications
DNA ligase is extensively used in recombinant DNA technology. The most frequently used enzymes are produced from the cloned genes of E. coli, Thermus aquaticus, and bacteriophage T4. Whenever one wishes to join two DNA fragments together, DNA ligase can be employed. For example, it is widely used in vector constructions to link DNA fragments to form chimeric molecules. DNA ligase is also used to attach duplex oligodeoxyribonucleotide linkers or adapters to a longer DNA to facilitate its further manipulation. A number of site-directed mutagenesis protocols depend on DNA ligase, in conjunction with a DNA polymerase, to form an intact, sequence-altered strand of DNA on an unaltered, circular template strand. Upon transfection and replication, progeny duplex molecules containing the mutation are produced. DNA ligase has often been used to join chemically synthesized, appropriately hybridized oligodeoxyribonucleotides for de novo gene synthesis. Because of the strict requirements for the structure and nature of its DNA substrate, the enzyme can also be used to determine whether a 3′-OH or 5′-PO4 exists at a DNA nick or if a gap is present in one strand of a DNA duplex. It can be used to quantify the number of nicks in a DNA molecule by using a radiolabeled adenylylated enzyme and measuring the amount of radioactive AMP released. When used to circularize short fragments, the enzyme has been used to study the stiffness, twisting, bending, and helical repeat of DNA. For example, because the ends of a fragment must be brought into proper alignment for effective ligation, one can learn about the number of base pairs per turn of the DNA. Similarly, DNA ligase can be used to study the ability of another protein to bend DNA on binding, because the bending can facilitate the ligation by bringing the ends of the duplex fragment into proximity. The enzyme has also proved useful for the synthesis of defined-sequence RNA by efficiently joining RNA strands on a DNA template (9). The ligase chain reaction, which depends on two pairs of oligodeoxyribonucleotides annealing to the top and bottom strands of a denatured target duplex at a given locus to form ligatable complexes, serves as an efficient DNA amplification method (10). The amplification is based on the principle that the products of the ligations on each strand can themselves serve as templates for further ligations of the excess starting oligonucleotides,
leading to an exponential increase in product on sequential heat denaturations and ligations. Use of a thermostable DNA ligase allows repeated denaturations of the nucleic acids without the need for addition of further enzyme. Because conditions can be chosen so that ligase strictly requires fully base-paired nucleotides at the nick, the ligase chain reaction can be used to detect mutations through the use of appropriately designed oligonucleotides (11). Oligonucleotides that hybridize to the target so that the nick is fully base-paired are amplified, whereas those that do not are unreactive. DNA ligase has also been used to enhance DNA sequencing by hybridization on microchips by joining fully base-paired oligonucleotides to form longer, detectable products (12). Although alternative schemes that do not require DNA ligase have been devised for accomplishing some of these functions, the enzyme remains a staple for nucleic acid manipulations.
References
1. I. R. Lehman (1974) Science 186, 790–797.
2. A. E. Tomkinson and D. S. Levin (1997) BioEssays 19, 893–901.
3. M. J. Engler and C. C. Richardson (1992) in The Enzymes, Vol. XV, pp. 3–29.
4. C. Cheng and S Shuman (1997) Nucleic Acids Res. 25, 1369–1374.
5. S. Shuman and B. Schwer (1995) Mol. Microbiol. 17, 405–410.
6. H. S. Subramanya, A. J. Doherty, S. R. Ashford, and D. Wigley (1996) Cell 85, 607–615.
7. V. Sriskanda and S. Shuman (1998) Nucleic Acids Res. 26, 525–531.
8. J. Sekiguchi and S. Shuman (1997) Biochemistry 36, 9073–9079.
9. M. J. Moore and P. A. Sharp (1992) Science 256, 992–997.
10. M. Wiedmann, W. J. Wilson, J. Czajka, J Luo, F. Barany, and C. A. Batt (1994) PCR Applications 3, S51–S64.
11. K. Abravaya, J. J. Carrino, S. Muldoon, and H. H. Lee (1995) Nucleic Acids Res. 23, 675–682.
12. S. Dubiley, E. Kirillov, Y. Lysov, and A. Mirzabekov (1997) Nucleic Acids Res. 25, 2259–
2265.
 الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
 اخر الاخبار
اخر الاخبار
اخبار العتبة العباسية المقدسة

الآخبار الصحية















 قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة
قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة "المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة
"المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة (نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)
(نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)


















