
Eukaryotic RNA Polymerases Consist of Many Subunits
 المؤلف:
JOCELYN E. KREBS, ELLIOTT S. GOLDSTEIN and STEPHEN T. KILPATRICK
المؤلف:
JOCELYN E. KREBS, ELLIOTT S. GOLDSTEIN and STEPHEN T. KILPATRICK
 المصدر:
LEWIN’S GENES XII
المصدر:
LEWIN’S GENES XII
 الجزء والصفحة:
الجزء والصفحة:
 7-5-2021
7-5-2021
 1838
1838
Eukaryotic RNA Polymerases Consist of Many Subunits
KEY CONCEPTS
- RNA polymerase I synthesizes rRNA in the nucleolus.
- RNA polymerase II synthesizes mRNA in the nucleoplasm.
- RNA polymerase III synthesizes small RNAs in the nucleoplasm.
- All eukaryotic RNA polymerases have about 12 subunits and are complexes of about 500 kD.
Some subunits are common to all three RNA polymerases.
- The largest subunit in RNA polymerase II has a carboxyterminal domain (CTD) consisting of multiple repeats of a heptamer sequence.
The three eukaryotic RNA polymerases have different locations in the nucleus that correspond with the different genes that they transcribe. The most prominent of the three with regard to activity is the enzyme RNA polymerase I, which resides in the nucleolus and is responsible for transcribing the genes coding for the 18S and 28S rRNA. It accounts for most cellular RNA synthesis (in terms of quantity).
The other major enzyme is RNA polymerase II, which is located in the nucleoplasm (i.e., the part of the nucleus excluding the nucleolus). It represents most of the remaining cellular activity and is responsible for synthesizing most of the heterogeneous nuclear RNA (hnRNA), the precursor for most mRNA and a lot more. The classical definition was that hnRNA includes everything but rRNA and tRNA in the nucleus (again, classically, mRNA is only found in the cytoplasm). With modern molecular tools, it is now possible to look a little closer at hnRNA. Researchers have found many lowabundance RNAs that are very important, plus many others that are just now beginning to be understood. mRNA is the least abundant of the three major RNAs, accounting for just 2% to 5% of the cytoplasmic RNA.
RNA polymerase III is a minor enzyme in terms of activity, but it produces a collection of stable, essential RNAs. This nucleoplasmic enzyme synthesizes the 5S rRNAs, tRNAs, and other small RNAs that constitute more than a quarter of the cytoplasmic RNAs.
All eukaryotic RNA polymerases are large proteins, functioning as complexes of approximately 500 kD. They typically have about 12 subunits. The purified enzymes can undertake template-dependent transcription of RNA, but are not able to initiate selectively at promoters. The general constitution of a eukaryotic RNA polymerase II enzyme as typified in Saccharomyces cerevisiae is illustrated in FIGURE 1. The two largest subunits are homologous to the β and β′ subunits of bacterial RNA polymerase.
Three of the remaining subunits are common to all the RNA polymerases; that is, they are also components of RNA polymerases I and III. Note that there is no subunit related to the bacterial sigma factor. Its function is contained in the basal transcription factors.
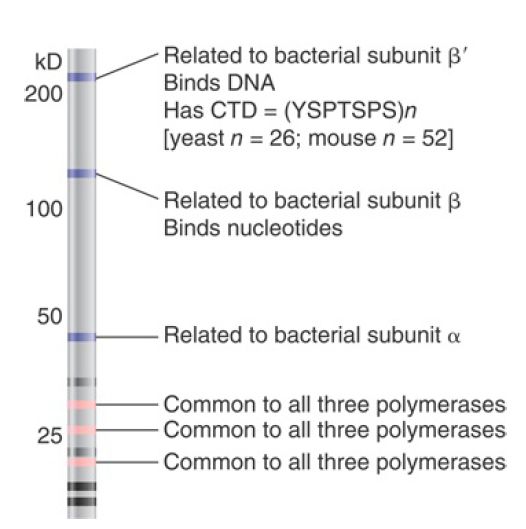
FIGURE 1 Some subunits are common to all classes of eukaryotic RNA polymerases and some are related to bacterial RNA polymerase. This drawing is a simulation of purified yeast RNA polymerase II run on an SDS gel to separate the subunits by size.
The largest subunit in RNA polymerase II has a carboxy-terminal domain (CTD), which consists of multiple repeats of a consensus sequence of seven amino acids. The sequence is unique to RNA polymerase II. Yeast has about 26 repeats and mammals have about 50. The number of repeats is important because deletions that remove (typically) more than half of the repeats are lethal. The CTD can be highly phosphorylated on serine or threonine residues.
The CTD is involved in regulating the initiation reaction , transcription elongation, and all aspects of mRNA processing, even export of mRNA to the cytoplasm. The RNA polymerases of mitochondria and chloroplasts are smaller, and they resemble bacterial RNA polymerase rather than any of the nuclear enzymes (because they evolved from eubacteria). Of course, the organelle genomes are much smaller; thus the resident polymerase needs to transcribe relatively few genes, and the control of transcription is likely to be very much simpler. These enzymes are more similar to bacteriophage enzymes that do not need to respond to a more complex environment.
A major practical distinction between the eukaryotic enzymes is drawn from their response to the bicyclic octapeptide α-amanitin (the toxic compound in Amanita mushroom species). In essentially all eukaryotic cells, the activity of RNA polymerase II is rapidly inhibited by low concentrations of α-amanitin (resulting in transcriptional shutdown leading to acute liver toxicity in Amanita poisoning). RNA polymerase I is not inhibited. The response of RNA polymerase III is less well conserved; in animal cells it is inhibited by high levels, but in yeast and insects it is not inhibited.
 الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
 اخر الاخبار
اخر الاخبار
اخبار العتبة العباسية المقدسة


