
Introduction to The Repair Systems
 المؤلف:
JOCELYN E. KREBS, ELLIOTT S. GOLDSTEIN and STEPHEN T. KILPATRICK
المؤلف:
JOCELYN E. KREBS, ELLIOTT S. GOLDSTEIN and STEPHEN T. KILPATRICK
 المصدر:
LEWIN’S GENES XII
المصدر:
LEWIN’S GENES XII
 الجزء والصفحة:
الجزء والصفحة:
 19-4-2021
19-4-2021
 2884
2884
Introduction to The Repair Systems
Any event that introduces a deviation from the usual double-helical structure of DNA is a threat to the genetic constitution of the cell.
Injury to DNA is minimized by systems that recognize and correct the damage. The repair systems are as complex as the replication apparatus itself, which indicates their importance for the survival of the cell. When a repair system reverses a change to DNA, there is no consequence. A mutation may result, though, when it fails to do so. The measured rate of mutation reflects a balance between the number of damaging events occurring in DNA and the number that have been corrected (or miscorrected).
Repair systems recognize a range of distortions in DNA as signals for action. The response to damage includes activation and recruitment of repair enzymes; modification of chromatin structure; activation of cell cycle checkpoints; and, in the event of insufficient repair in multicellular organisms, apoptosis. The importance of DNA repair in eukaryotes is indicated by the identification of more than 130 repair genes in the human genome. As summarized in FIGURE 1, we can divide the repair systems into several general types:
- Some enzymes directly reverse specific sorts of damage to DNA.
- Pathways exist for base excision repair, nucleotide excision repair, and mismatch repair, all of which function by removing damaged/mispaired regions and synthesizing new DNA using the intact strand as a template.
- Some systems function by using recombination to retrieve an undamaged copy that is then used to replace a damaged duplex sequence.
- The nonhomologous end-joining pathway rejoins broken doublestrand ends.
- Translesion or error-prone DNA polymerases can bypass certain damage or synthesize stretches of replacement DNA that may contain additional errors.
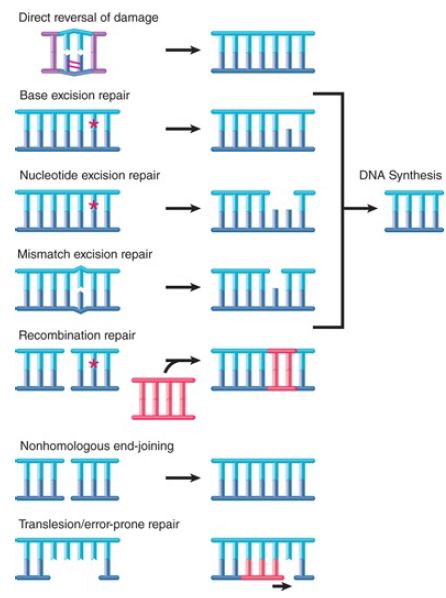
FIGURE .1 Repair systems can be classified into pathways that use different mechanisms to reverse or bypass damage to DNA.
Direct repair is rare and involves the reversal or simple removal of the damage. One good example is photoreactivation of pyrimidine dimers, in which inappropriate covalent bonds between adjacent bases are reversed by a light-dependent enzyme.
Several pathways of excision repair entail removal of incorrect or damaged sequences followed by repair synthesis. Excision repair pathways are initiated by recognition enzymes that see an actual damaged base or a change in the spatial path of DNA. FIGURE .2 summarizes the main events in a generic excision repair pathway. Some excision repair pathways recognize general damage to DNA; others act upon specific types of base damage. A single cell type usually has multiple excision repair systems.
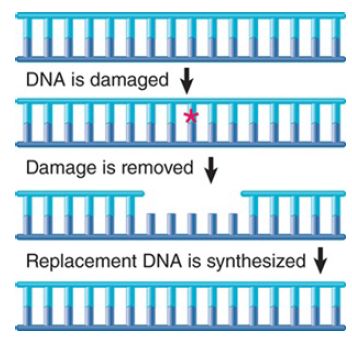
FIGURE .2 Excision repair directly replaces damaged DNA and then resynthesizes a replacement stretch for the damaged strand.
Mismatches between the strands of DNA are one of the major targets for excision repair systems. Mismatch repair (MMR) is accomplished by scrutinizing DNA for apposed bases that do not pair properly. This system also recognizes insertion/deletion loops in which sequences present in one strand that are absent in the complementary strand are looped out. Mismatches and insertion/deletion loops that arise during replication are corrected by distinguishing between the “new” and “old” strands and preferentially correcting the sequence of the newly synthesized strand. Other systems deal with mismatches generated by base conversions, such as the result of deamination.
The two major excision repair pathways, in addition to mismatch repair, are as follows:
- Base excision repair (BER) systems directly remove the damaged base and replace it in DNA. A good example is uracil-DNA glycosylase (UDG; also known as uracil N-glycosylase, UNG), which removes uracils that are mispaired with guanines .
- Nucleotide excision repair (NER) systems excise a sequence that includes the damaged base(s); a new stretch of DNA is then synthesized to replace the excised material.
In contrast to excision repair mechanisms, recombination-repair systems handle situations in which damage remains in a daughter molecule and replication has been forced to bypass the site, which typically creates a gap in the daughter strand. A retrieval system uses recombination to obtain another copy of the sequence from an undamaged source; the copy is then used to repair the gap.
A major feature in recombination and repair is the need to handle double-strand breaks (DSBs), which can arise from a variety of mechanisms. DSBs are intentionally created to initiate crossovers during homologous recombination in meiosis. They can also be created by problems in replication, when they may trigger the use of recombination-repair systems. DSBs can also be created by environmental damage (e.g., by radiation damage), intrinsic damage (reactive oxygen species resulting from cellular metabolism), or can be the result from the shortening of telomeres to expose nontelomeric chromosome ends. In all of these events, DSBs can cause mutations, including loss of large chromosomal regions. DSBs can be repaired via recombination-repair using homologous sequences or by joining together nonhomologous DNA ends.
Mutations that affect the ability of Escherichia coli cells to engage in DNA repair fall into groups that correspond to several repair pathways (not necessarily all independent). The major known pathways are the uvr excision repair system, the methyl-directed mut mismatch repair system, and the recB and recF recombination and recombination-repair pathways. The enzyme activities associated with these systems are endonucleases and exonucleases (important in removing damaged DNA); resolvases (endonucleases that act specifically on recombinant junctions); helicases to unwind DNA; and DNA polymerases to synthesize new DNA. Some of these enzyme activities are unique to particular repair pathways, whereas others participate in multiple pathways.
The replication apparatus devotes a lot of attention to quality control. DNA polymerases use proofreading to check the daughter strand sequence and to remove errors. Some of the repair systems are less accurate when they synthesize DNA to replace damaged material. For this reason, these systems have been known historically as error-prone systems.
 الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
 اخر الاخبار
اخر الاخبار
اخبار العتبة العباسية المقدسة


