

النبات

مواضيع عامة في علم النبات

الجذور - السيقان - الأوراق

النباتات الوعائية واللاوعائية

البذور (مغطاة البذور - عاريات البذور)

الطحالب

النباتات الطبية


الحيوان

مواضيع عامة في علم الحيوان

علم التشريح

التنوع الإحيائي

البايلوجيا الخلوية


الأحياء المجهرية

البكتيريا

الفطريات

الطفيليات

الفايروسات


علم الأمراض

الاورام

الامراض الوراثية

الامراض المناعية

الامراض المدارية

اضطرابات الدورة الدموية

مواضيع عامة في علم الامراض

الحشرات


التقانة الإحيائية

مواضيع عامة في التقانة الإحيائية


التقنية الحيوية المكروبية

التقنية الحيوية والميكروبات

الفعاليات الحيوية

وراثة الاحياء المجهرية

تصنيف الاحياء المجهرية

الاحياء المجهرية في الطبيعة

أيض الاجهاد

التقنية الحيوية والبيئة

التقنية الحيوية والطب

التقنية الحيوية والزراعة

التقنية الحيوية والصناعة

التقنية الحيوية والطاقة

البحار والطحالب الصغيرة

عزل البروتين

هندسة الجينات


التقنية الحياتية النانوية

مفاهيم التقنية الحيوية النانوية

التراكيب النانوية والمجاهر المستخدمة في رؤيتها

تصنيع وتخليق المواد النانوية

تطبيقات التقنية النانوية والحيوية النانوية

الرقائق والمتحسسات الحيوية

المصفوفات المجهرية وحاسوب الدنا

اللقاحات

البيئة والتلوث


علم الأجنة

اعضاء التكاثر وتشكل الاعراس

الاخصاب

التشطر

العصيبة وتشكل الجسيدات

تشكل اللواحق الجنينية

تكون المعيدة وظهور الطبقات الجنينية

مقدمة لعلم الاجنة


الأحياء الجزيئي

مواضيع عامة في الاحياء الجزيئي


علم وظائف الأعضاء


الغدد

مواضيع عامة في الغدد

الغدد الصم و هرموناتها

الجسم تحت السريري

الغدة النخامية

الغدة الكظرية

الغدة التناسلية

الغدة الدرقية والجار الدرقية

الغدة البنكرياسية

الغدة الصنوبرية

مواضيع عامة في علم وظائف الاعضاء

الخلية الحيوانية

الجهاز العصبي

أعضاء الحس

الجهاز العضلي

السوائل الجسمية

الجهاز الدوري والليمف

الجهاز التنفسي

الجهاز الهضمي

الجهاز البولي


المضادات الميكروبية

مواضيع عامة في المضادات الميكروبية

مضادات البكتيريا

مضادات الفطريات

مضادات الطفيليات

مضادات الفايروسات

علم الخلية

الوراثة

الأحياء العامة

المناعة

التحليلات المرضية

الكيمياء الحيوية

مواضيع متنوعة أخرى

الانزيمات
The Bacterial Genome Is a Nucleoid with Dynamic Structural Properties
المؤلف:
JOCELYN E. KREBS, ELLIOTT S. GOLDSTEIN and STEPHEN T. KILPATRICK
المصدر:
LEWIN’S GENES XII
الجزء والصفحة:
21-3-2021
3954
The Bacterial Genome Is a Nucleoid with Dynamic Structural Properties
KEY CONCEPTS
-The bacterial nucleoid is organized as multiple loops compacted by nucleoid-associated proteins such as HNS and HU.
-Nucleoid-associated proteins are typically small, abundant, DNA-binding proteins that function in nucleoid architecture, domain topology, and gene regulation.
-Bacterial condensin complexes (SMC-ScpAB or MukBEF) function in chromosome structure and segregation.
Although bacteria do not display structures with the distinct morphological features of eukaryotic chromosomes, their genomes nonetheless are organized into definite substructures within the cell.
We can see the genetic material as a fairly compact clump (or series of clumps) that occupies about a third of the volume of the cell. FIGURE 1 displays a thin section through a bacterium in which this nucleoid is evident.

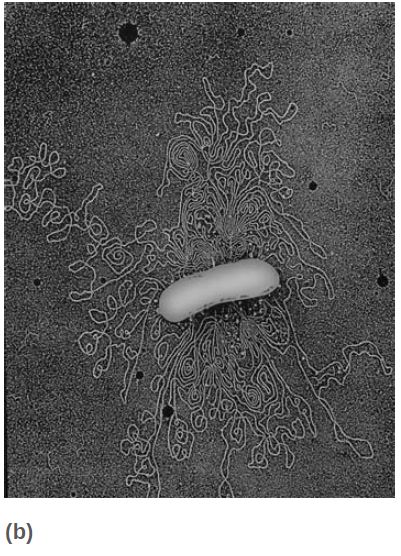
FIGURE 1 (a) A thin section shows the bacterial nucleoid as a compact mass in the center of the cell. (b) The nucleoid spills out of a lysed E. coli cell in the form of loops of a fiber.
(a) Photo courtesy of the Molecular and Cell Biology Instructional Laboratory Program, University of California, Berkeley.
(b) © Dr. Gopal Murti/Science Source.
When E. coli cells are lysed, fibers are released in the form of loops attached to the broken envelope of the cell, as shown in Figure 1b. The DNA of these loops is not found in the extended form of a free duplex, but instead is compacted by association with proteins.
Increasing numbers of nucleoid-associated proteins (NAPs) that resemble eukaryotic chromosomal proteins have been isolated in archaea and bacteria. Exactly what constitutes a NAP is vague, because some of them might contribute to multiple genetic functions. As a group, NAPs are emerging as antagonistic regulators of gene activity and nucleoid structure. In the gramnegative bacteria, researchers have characterized as many as 12 different NAPs, some of them depicted in FIGURE 2.
Most NAPs have DNA-binding activities that can affect the spatial arrangement of DNA through bending, wrapping, or bridging.
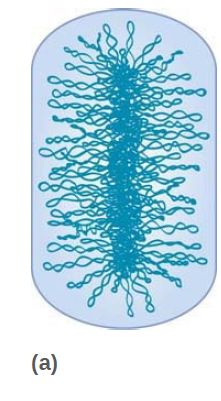
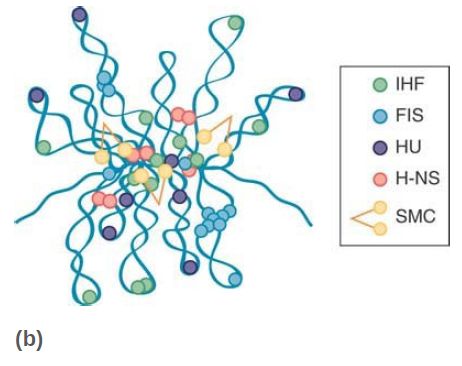
FIGURE 2 Topological organization of the bacterial chromosome. (a) Schematic representation of the bottlebrush model of the nucleoid. This diagram depicts the interwound supercoiled loops emanating from a dense core. The topologically isolated domains are on average 10 kb and therefore are likely to encompass several branched plectonemic loops. (b) Schematic representation of the small nucleoid-associated proteins and the structural maintenance of chromosome (SMC) complexes. These proteins introduce DNA bends and also function in bridging chromosomal loci.
FIGURE 3 summarizes how NAPs vary in their function and expression patterns as cells progress through growth phases. The dynamics of individual NAPs and their interactions with one another are becoming increasingly more clear despite the complexity of their multifaceted effects on nucleoid structure and function.

FIGURE 3 Growth phase and elements that affect nucleoid structure. A typical growth curve for E. coli growing in batch culture begins with a lag phase followed by the log phase of exponential growth and, finally, stationary phase (when the cells stop growing). Important nucleoid-associated proteins are expressed at different times during the growth curve, as indicated. In addition, there are significant changes in DNA topology: DNA is negatively supercoiled (SC) in log phase cells, whereas it is more relaxed (R) in lag phase and stationary phase cells.
Protein H-NS (histone-like, nucleoid-structuring protein) has a preference for AT-rich DNA and can form DNA-H-NS-DNA bridges, allowing this NAP to simultaneously influence gene promoter activity and nucleoid structure. H-NS is expressed throughout all growth phases. Its interactions with other expression-modulating proteins likely contribute to the ability of H-NS to silence hundreds of genes and form boundaries of microdomains. Recent advances in chromosome conformation capture (C3; also see the chapter titled Chromatin) and high-resolution fluorescence imaging suggest that H-NS might mediate the colocalization of many H-NS-binding sites into two foci. These have been proposed to represent each of two replichores, the left and right arms of the circular genome that are replicated by the bidirectional movement forks from the origin.
Protein HU has two subunits: homodimers or heterodimers of HUα and HUβ. They bend or wrap DNA and play a role in DNA flexibility.
These histone-like proteins bind nonspecifically to multiple sites with some preference for distorted DNA regions such as bends, forks, four-way junctions, nicks, or overhangs. Consequently, they are implicated as architectural factors affecting various functions in DNA metabolism.
Other NAPs, such as IHF, Dps, and bacterial condensins, also appear to have multiple or overlapping roles in nucleoid architecture and core genetic processes. One of these is the integration host factor (IHF), first identified as a bacteriophage lambda cofactor for site-specific integration. IHF has since been found to bend DNA and induce U-turns and influence global transcription, not unlike a general transcription factor. The ability of IHF to alter local DNA structure through U-turn formation appears to be a defining feature of its mode of action in replication, phage integration, transposition, and transcription. Another well-characterized and interesting NAP is the DNA protection during starvation protein (Dps). Dps is expressed in the stationary phase and in oxidatively stressed cells, likely functioning to limit DNA damage. MukB and its homologs are chromosome structural maintenance proteins that are now recognized as components of bacterial condensin complexes.
Similar to eukaryotic condensins in structure and function, they regulate chromosome condensation and are required for proper segregation of chromosomes during cell division. Genetic evidence establishes a role for these complexes (MukBEF or SMC-ScpAB) in DNA topology and domain delineation.
As a group, the NAP proteins and their expression patterns point to an integrating principle whereby nucleoid structure and gene expression are comodulated during cell growth and reproduction in an environmentally responsive manner. How these packaging functions are coupled to gene positioning and promoter functions to affect bacterial fitness and to what extent such an integrated system imposes evolutionary constraints for bacterial fitness are among the key questions in bacterial functional genomics.
 الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
 اخر الاخبار
اخر الاخبار
اخبار العتبة العباسية المقدسة

الآخبار الصحية















 قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة
قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة "المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة
"المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة (نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)
(نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)


















