

النبات

مواضيع عامة في علم النبات

الجذور - السيقان - الأوراق

النباتات الوعائية واللاوعائية

البذور (مغطاة البذور - عاريات البذور)

الطحالب

النباتات الطبية


الحيوان

مواضيع عامة في علم الحيوان

علم التشريح

التنوع الإحيائي

البايلوجيا الخلوية


الأحياء المجهرية

البكتيريا

الفطريات

الطفيليات

الفايروسات


علم الأمراض

الاورام

الامراض الوراثية

الامراض المناعية

الامراض المدارية

اضطرابات الدورة الدموية

مواضيع عامة في علم الامراض

الحشرات


التقانة الإحيائية

مواضيع عامة في التقانة الإحيائية


التقنية الحيوية المكروبية

التقنية الحيوية والميكروبات

الفعاليات الحيوية

وراثة الاحياء المجهرية

تصنيف الاحياء المجهرية

الاحياء المجهرية في الطبيعة

أيض الاجهاد

التقنية الحيوية والبيئة

التقنية الحيوية والطب

التقنية الحيوية والزراعة

التقنية الحيوية والصناعة

التقنية الحيوية والطاقة

البحار والطحالب الصغيرة

عزل البروتين

هندسة الجينات


التقنية الحياتية النانوية

مفاهيم التقنية الحيوية النانوية

التراكيب النانوية والمجاهر المستخدمة في رؤيتها

تصنيع وتخليق المواد النانوية

تطبيقات التقنية النانوية والحيوية النانوية

الرقائق والمتحسسات الحيوية

المصفوفات المجهرية وحاسوب الدنا

اللقاحات

البيئة والتلوث


علم الأجنة

اعضاء التكاثر وتشكل الاعراس

الاخصاب

التشطر

العصيبة وتشكل الجسيدات

تشكل اللواحق الجنينية

تكون المعيدة وظهور الطبقات الجنينية

مقدمة لعلم الاجنة


الأحياء الجزيئي

مواضيع عامة في الاحياء الجزيئي


علم وظائف الأعضاء


الغدد

مواضيع عامة في الغدد

الغدد الصم و هرموناتها

الجسم تحت السريري

الغدة النخامية

الغدة الكظرية

الغدة التناسلية

الغدة الدرقية والجار الدرقية

الغدة البنكرياسية

الغدة الصنوبرية

مواضيع عامة في علم وظائف الاعضاء

الخلية الحيوانية

الجهاز العصبي

أعضاء الحس

الجهاز العضلي

السوائل الجسمية

الجهاز الدوري والليمف

الجهاز التنفسي

الجهاز الهضمي

الجهاز البولي


المضادات الميكروبية

مواضيع عامة في المضادات الميكروبية

مضادات البكتيريا

مضادات الفطريات

مضادات الطفيليات

مضادات الفايروسات

علم الخلية

الوراثة

الأحياء العامة

المناعة

التحليلات المرضية

الكيمياء الحيوية

مواضيع متنوعة أخرى

الانزيمات
The Rate of Neutral Substitution Can Be Measured from Divergence of Repeated Sequences
المؤلف:
JOCELYN E. KREBS, ELLIOTT S. GOLDSTEIN and STEPHEN T. KILPATRICK
المصدر:
LEWIN’S GENES XII
الجزء والصفحة:
15-3-2021
2393
The Rate of Neutral Substitution Can Be Measured from Divergence of Repeated Sequences
KEY CONCEPT
-The rate of substitution per year at neutral sites is greater in the mouse genome than in the human genome, probably because of a higher mutation rate.
We can make the best estimate of the rate of substitution at neutral sites by examining sequences that do not encode polypeptide. (We use the term neutral here rather than synonymous because there is no coding potential.) An informative comparison can be made by comparing the members of a common repetitive family in the human and mouse genomes.
The principle of the analysis is summarized in FIGURE 1. We begin with a family of related sequences that have evolved by duplication and substitution from an original ancestral sequence.
We assume that the ancestral sequence can be deduced by taking the base that is most common at each position. Then we can calculate the divergence of each individual family member as the
proportion of bases that differ from the deduced ancestral sequence. In this example, individual members vary from 0.13 to 0.18 divergence and the average is 0.16.
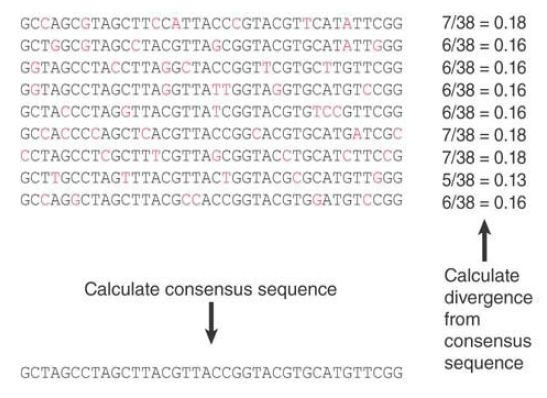
FIGURE 1. An ancestral consensus sequence for a family is calculated by taking the most common base at each position. The divergence of each existing current member of the family is calculated as the proportion of bases at which it differs from the ancestral sequence.
One family used for this analysis in the human and mouse genomes derives from a sequence that is thought to have ceased to be functional at about the time of the common ancestor between humans and rodents . This means that it has been diverging under limited selective pressure for the same length of time in both species. Its average divergence in humans is about
0.17 substitutions per site, corresponding to a rate of 2.2 × 10−9 substitutions per base per year over the 75 million years since the separation. However, in the mouse genome, neutral substitutions have occurred at twice this rate, corresponding to 0.34 substitutions per site in the family, or a rate of 4.5 × 10-8 . Note, however, that if we calculated the rate per generation instead of per year, it would be greater in humans than in the mouse (2.2 × 10-9 as opposed to 10 ).
These figures probably underestimate the rate of substitution in the mouse; at the time of divergence, the rates in both lineages would have been the same and the difference must have evolved since then. The current rate of neutral substitution per year in the mouse is probably two to three times greater than the historical average.
At first glance, these rates would seem to reflect the balance between the occurrence of mutations (which can be higher in species with higher metabolic rates, like the mouse) and the loss of them due to genetic drift, which is largely a function of population size, because genetic drift is a type of “sampling error” where allele frequencies fluctuate more widely in smaller populations. In addition to eliminating neutral alleles more quickly, smaller population sizes also allow faster fixation and loss of neutral alleles. Rodent species tend to have short generation times (allowing more opportunities for substitutions per year), but species with short generation times also tend to have larger population sizes, so the effects of more substitutions per year but less fixation of neutral alleles would cancel each other out. The higher substitution rate in mice is probably due primarily to a higher mutation rate.
Comparing the mouse and human genomes allows us to assess whether syntenic (homologous) regions show signs of conservation or have differed at the rate predicted from accumulation of neutral substitutions. The proportion of sites that show signs of selection is about 5%. This is much higher than the proportion found in exons (about 1%). This observation implies that the genome includes many more stretches whose sequence is important for functions other than encoding RNA. Known regulatory elements are likely to comprise only a small part of this proportion. This number also suggests that most (i.e., the rest) of the genome sequences do not
have any function that depends on the exact sequence.
 الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
 اخر الاخبار
اخر الاخبار
اخبار العتبة العباسية المقدسة

الآخبار الصحية















 قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة
قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة "المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة
"المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة (نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)
(نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)


















