

النبات

مواضيع عامة في علم النبات

الجذور - السيقان - الأوراق

النباتات الوعائية واللاوعائية

البذور (مغطاة البذور - عاريات البذور)

الطحالب

النباتات الطبية


الحيوان

مواضيع عامة في علم الحيوان

علم التشريح

التنوع الإحيائي

البايلوجيا الخلوية


الأحياء المجهرية

البكتيريا

الفطريات

الطفيليات

الفايروسات


علم الأمراض

الاورام

الامراض الوراثية

الامراض المناعية

الامراض المدارية

اضطرابات الدورة الدموية

مواضيع عامة في علم الامراض

الحشرات


التقانة الإحيائية

مواضيع عامة في التقانة الإحيائية


التقنية الحيوية المكروبية

التقنية الحيوية والميكروبات

الفعاليات الحيوية

وراثة الاحياء المجهرية

تصنيف الاحياء المجهرية

الاحياء المجهرية في الطبيعة

أيض الاجهاد

التقنية الحيوية والبيئة

التقنية الحيوية والطب

التقنية الحيوية والزراعة

التقنية الحيوية والصناعة

التقنية الحيوية والطاقة

البحار والطحالب الصغيرة

عزل البروتين

هندسة الجينات


التقنية الحياتية النانوية

مفاهيم التقنية الحيوية النانوية

التراكيب النانوية والمجاهر المستخدمة في رؤيتها

تصنيع وتخليق المواد النانوية

تطبيقات التقنية النانوية والحيوية النانوية

الرقائق والمتحسسات الحيوية

المصفوفات المجهرية وحاسوب الدنا

اللقاحات

البيئة والتلوث


علم الأجنة

اعضاء التكاثر وتشكل الاعراس

الاخصاب

التشطر

العصيبة وتشكل الجسيدات

تشكل اللواحق الجنينية

تكون المعيدة وظهور الطبقات الجنينية

مقدمة لعلم الاجنة


الأحياء الجزيئي

مواضيع عامة في الاحياء الجزيئي


علم وظائف الأعضاء


الغدد

مواضيع عامة في الغدد

الغدد الصم و هرموناتها

الجسم تحت السريري

الغدة النخامية

الغدة الكظرية

الغدة التناسلية

الغدة الدرقية والجار الدرقية

الغدة البنكرياسية

الغدة الصنوبرية

مواضيع عامة في علم وظائف الاعضاء

الخلية الحيوانية

الجهاز العصبي

أعضاء الحس

الجهاز العضلي

السوائل الجسمية

الجهاز الدوري والليمف

الجهاز التنفسي

الجهاز الهضمي

الجهاز البولي


المضادات الميكروبية

مواضيع عامة في المضادات الميكروبية

مضادات البكتيريا

مضادات الفطريات

مضادات الطفيليات

مضادات الفايروسات

علم الخلية

الوراثة

الأحياء العامة

المناعة

التحليلات المرضية

الكيمياء الحيوية

مواضيع متنوعة أخرى

الانزيمات
The Human Genome Has Fewer Genes Than Originally Expected
المؤلف:
JOCELYN E. KREBS, ELLIOTT S. GOLDSTEIN and STEPHEN T. KILPATRICK
المصدر:
LEWIN’S GENES XII
الجزء والصفحة:
14-3-2021
2763
The Human Genome Has Fewer Genes Than Originally Expected
KEY CONCEPTS
-Only 1% of the human genome consists of exons.
-The exons comprise about 5% of each gene, so genes (exons plus introns) comprise about 25% of the genome.
-The human genome has about 20,000 genes.
-Roughly 60% of human genes are alternatively spliced.
-Up to 80% of the alternative splices change protein sequence, so the human proteome has 50,000 to 60,000 members.
The human genome was the first vertebrate genome to be sequenced. This massive task has revealed a wealth of information about the genetic makeup of our species and about the evolution of genomes in general. Our understanding is deepened further by the ability to compare the human genome sequence with other sequenced vertebrate genomes.
Mammal genomes generally fall into a narrow size range, averaging about 3 × 109 bp . The mouse genome is about 14% smaller than the human genome, probably because it has had a higher rate of deletion. The genomes contain similar gene families and genes, with most genes having an ortholog in the other genome but with differences in the number of members of a family, especially in those cases for which the functions are specific to the species . Originally estimated to have about 30,000 genes, the mouse genome is now estimated to have more protein-coding genes than the human genome does, about 25,000.
FIGURE 1. plots the distribution of the mouse genes. The 25,000 protein-coding genes are accompanied by about 3,000 genes representing RNAs that do not encode proteins; these are generally small (aside from the ribosomal RNAs). Almost half of these genes encode transfer RNAs. In addition to the functional genes, about 1,200 pseudogenes have been identified.
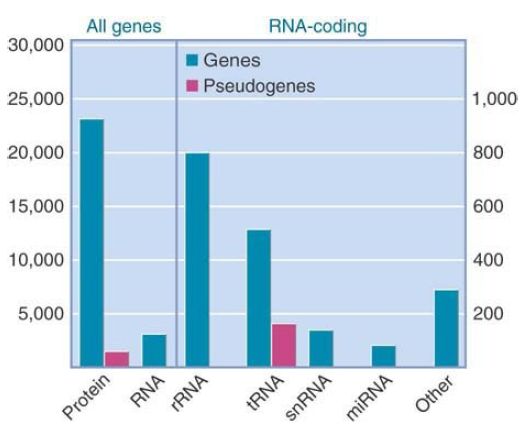
FIGURE 5.8 The mouse genome has about 25,000 protein-coding genes, which include about 1,200 pseudogenes. There are about 3,000 RNA-coding genes.
The haploid human genome contains 22 autosomes plus the X and Y chromosomes. The chromosomes range in size from 45 to 279 Mb, making a total genome size of 3,235 Mb (about 3.2 × 109 bp). On the basis of chromosome structure, the genome can be divided into regions of euchromatin (containing many functional genes) and heterochromatin, with a much lower density of functional genes . The euchromatin comprises the majority of the genome, about 2.9 × 109 bp. The identified genome sequence represents more than 90% of the euchromatin. In addition to providing information on the genetic content of the genome, the sequence also identifies features that may be of structural importance.
FIGURE 2. shows that a very small proportion (about 1%) of the human genome is accounted for by the exons that actually encode polypeptides. The introns that constitute the remaining sequences of protein-coding genes bring the total of DNA involved with producing proteins to about 25%. As shown in FIGURE 3, the average human gene is 27 kb long with nine exons that include a total coding sequence of 1,340 bp. Therefore, the average coding sequence is only 5% of the length of an average protein-coding gene.
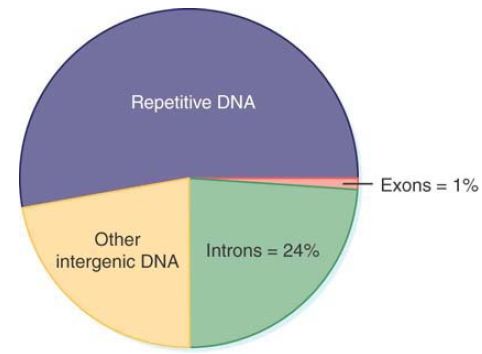
FIGURE 2. Genes occupy 25% of the human genome, but protein-coding sequences are only a small part of this fraction.
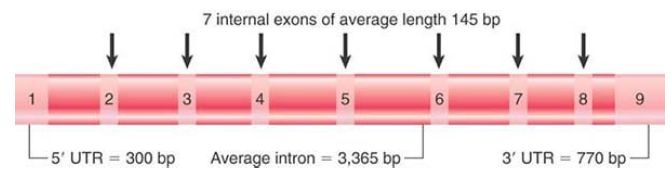
FIGURE 3. The average human gene is 27 kb long and has 9 exons usually comprising 2 longer exons at each end and 7 internal exons. The UTRs in the terminal exons are the untranslated (noncoding) regions at each end of the gene. (This is based on the average. Some genes are extremely long, which makes the median length 14 kb with 7 exons.)
Two independent sequencing efforts for the human genome produced estimates of 30,000 and 40,000 genes, respectively.
One measure of the accuracy of the analyses is whether they identify the same genes. The surprising answer is that the overlap between the two sets of genes is only about 50%, as summarized in FIGURE 4. An earlier analysis of the human gene set based on RNA transcripts had identified about 11,000 genes, almost all of which are present in both the large human gene sets, and which account for the major part of the overlap between them. So there is no question about the authenticity of half of each human gene set, but we have yet to establish the relationship between the other half of each set. The discrepancies illustrate the pitfalls of large-scale sequence analysis! As the sequence is analyzed further (and as other genomes are sequenced with which it can be compared), the number of actual genes has declined, and is now estimated to be about 20,000.
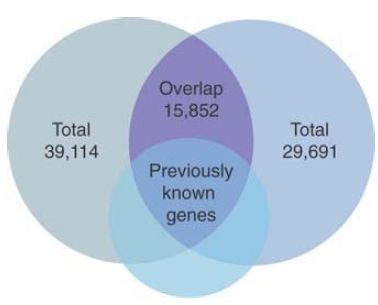
FIGURE 4. The two sets of genes identified in the human genome overlap only partially, as shown in the two large upper circles. However, they include almost all previously known genes, as shown by the overlap with the smaller, lower circle.
By any measure, the total human gene number is much smaller than was originally estimated—most estimates before the genome was sequenced were about 100,000. This represents a relatively small increase over the gene number of fruit flies and nematode worms (recent work suggests as many as 17,000 and 21,700, respectively), not to mention the plants Arabidopsis (25,000) and rice (32,000). However, we should not be particularly surprised by the notion that it does not take a great number of additional genes to make a more complex organism. The difference in DNA sequences between the human and chimpanzee genomes is extremely small (there is 98.5% similarity), so it is clear that the functions and interactions between a similar set of genes can produce different results. The functions of specific groups of genes can be especially important because detailed comparisons of orthologous genes in humans and chimpanzees suggest that there has been rapid evolution of certain classes of genes, including some involved in early development, olfaction, and hearing—all functions that are relatively specialized in these species.
The number of protein-coding genes is less than the number of potential polypeptides because of mechanisms such as alternative splicing, alternate promoter selection, and alternate poly(A) site selection that can result in several polypeptides from the same gene (see the RNA Splicing and Processing chapter). The extent of alternative splicing is greater in humans than in flies or worms; it affects more than 60% of the genes (perhaps more than 90%), so the increase in size of the human proteome relative to that of the other eukaryotes might be larger than the increase in the number of genes. A sample of genes from two chromosomes suggests that the proportion of the alternative splices that actually result in changes in the polypeptide sequence is about 80%. If this occurs genome-wide, the size of the proteome could be 50,000 to 60,000 members.
However, in terms of the diversity of the number of gene families, the discrepancy between humans and the other eukaryotes might not be so great. Many of the human genes belong to gene families. An analysis of more than 20,000 genes identified 3,500 unique genes and 10,300 gene pairs.
 الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
 اخر الاخبار
اخر الاخبار
اخبار العتبة العباسية المقدسة

الآخبار الصحية















 قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة
قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة "المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة
"المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة (نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)
(نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)


















