

النبات

مواضيع عامة في علم النبات

الجذور - السيقان - الأوراق

النباتات الوعائية واللاوعائية

البذور (مغطاة البذور - عاريات البذور)

الطحالب

النباتات الطبية


الحيوان

مواضيع عامة في علم الحيوان

علم التشريح

التنوع الإحيائي

البايلوجيا الخلوية


الأحياء المجهرية

البكتيريا

الفطريات

الطفيليات

الفايروسات


علم الأمراض

الاورام

الامراض الوراثية

الامراض المناعية

الامراض المدارية

اضطرابات الدورة الدموية

مواضيع عامة في علم الامراض

الحشرات


التقانة الإحيائية

مواضيع عامة في التقانة الإحيائية


التقنية الحيوية المكروبية

التقنية الحيوية والميكروبات

الفعاليات الحيوية

وراثة الاحياء المجهرية

تصنيف الاحياء المجهرية

الاحياء المجهرية في الطبيعة

أيض الاجهاد

التقنية الحيوية والبيئة

التقنية الحيوية والطب

التقنية الحيوية والزراعة

التقنية الحيوية والصناعة

التقنية الحيوية والطاقة

البحار والطحالب الصغيرة

عزل البروتين

هندسة الجينات


التقنية الحياتية النانوية

مفاهيم التقنية الحيوية النانوية

التراكيب النانوية والمجاهر المستخدمة في رؤيتها

تصنيع وتخليق المواد النانوية

تطبيقات التقنية النانوية والحيوية النانوية

الرقائق والمتحسسات الحيوية

المصفوفات المجهرية وحاسوب الدنا

اللقاحات

البيئة والتلوث


علم الأجنة

اعضاء التكاثر وتشكل الاعراس

الاخصاب

التشطر

العصيبة وتشكل الجسيدات

تشكل اللواحق الجنينية

تكون المعيدة وظهور الطبقات الجنينية

مقدمة لعلم الاجنة


الأحياء الجزيئي

مواضيع عامة في الاحياء الجزيئي


علم وظائف الأعضاء


الغدد

مواضيع عامة في الغدد

الغدد الصم و هرموناتها

الجسم تحت السريري

الغدة النخامية

الغدة الكظرية

الغدة التناسلية

الغدة الدرقية والجار الدرقية

الغدة البنكرياسية

الغدة الصنوبرية

مواضيع عامة في علم وظائف الاعضاء

الخلية الحيوانية

الجهاز العصبي

أعضاء الحس

الجهاز العضلي

السوائل الجسمية

الجهاز الدوري والليمف

الجهاز التنفسي

الجهاز الهضمي

الجهاز البولي


المضادات الميكروبية

مواضيع عامة في المضادات الميكروبية

مضادات البكتيريا

مضادات الفطريات

مضادات الطفيليات

مضادات الفايروسات

علم الخلية

الوراثة

الأحياء العامة

المناعة

التحليلات المرضية

الكيمياء الحيوية

مواضيع متنوعة أخرى

الانزيمات
Isolation of DNA
المؤلف:
John M Walker and Ralph Rapley
المصدر:
Molecular Biology and Biotechnology 5th Edition
الجزء والصفحة:
6-11-2020
2040
Isolation of DNA
The use of DNA for analysis or manipulation usually requires that it is isolated and purified to a certain extent. DNA is recovered from cells by the gentlest possible method of cell rupture to prevent the DNA from fragmenting by mechanical shearing. This is usually in the presence of EDTA which chelate the Mg2+ ions needed for enzymes that degrade DNA termed DNase. Ideally, cell walls, if present, should be digested enzymatically (e.g. lysozyme treatment of bacteria) and the cell membrane should be solubilised using detergent. If physical disruption is necessary, it should be kept to a minimum and should involve cutting or squashing of cells, rather than the use of shear forces. Cell disruption (and most subsequent steps) should be performed at 4 ºC, using glassware and solutions which have been autoclaved to destroy DNase activity (Figure 1).
After release of nucleic acids from the cells, RNA can be removed by treatment with ribonuclease (RNase) which has been heat treated to inactivate any DNase contaminants; RNase is relatively stable to heat as a result of its disulfide bonds, which ensure rapid renaturation of the molecule on cooling. The other major contaminant, protein, is removed by shaking the solution gently with water-saturated phenol or with a
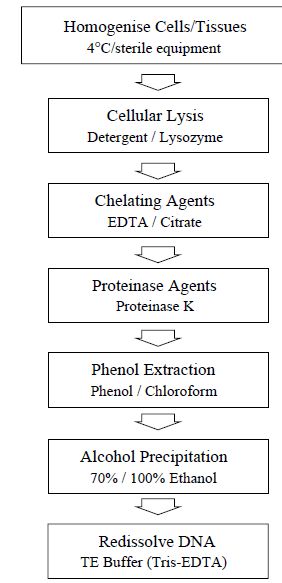
Figure 1: Flow diagram of the main steps involved in the extraction of DNA.
phenol–chloroform mixture, either of which will denature proteins but not nucleic acids. Centrifugation of the emulsion formed by this mixing produces a lower, organic phase, separated from the upper, aqueous phase by an interface of denatured protein. The aqueous solution is recovered and deproteinised repeatedly, until no more material is seen at the interface. Finally, the deproteinised DNA preparation is mixed with two volumes of absolute ethanol and the DNA allowed to precipitate out of solution in a freezer. After centrifugation, the DNA pellet is redissolved in a buffer containing EDTA to inactivate any DNases present. This solution can be stored at 4 ºC for at least 1 month. DNA solutions can be stored frozen, although repeated freezing and thawing tend to damage long DNA molecules by shearing. The procedure described above is suitable for total cellular DNA. If the DNA from a specific organelle or viral particle is needed, it is best to isolate the organelle or virus before extracting its DNA, since the
recovery of a particular type of DNA from a mixture is usually rather difficult. Where a high degree of purity is required, DNA may be subjected to density gradient ultracentrifugation through caesium chloride, which is particularly useful for the preparation of plasmid DNA. It is possible to check the integrity of the DNA by agarose gel electrophoresis and determine the concentration of the DNA by using the fact that
1 absorbance unit equates to 50 mgml-1 of DNA and so
50 * A260 = concentration of DNA sample (μgml-1)
Contaminants may also be identified by scanning UV spectrophotometry from 200 to 300 nm. A ratio of 260 nm:280nm of approximately 1.8 indicates that the sample is free of protein contamination, which absorbs strongly at 280 nm.
 الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
 اخر الاخبار
اخر الاخبار
اخبار العتبة العباسية المقدسة

الآخبار الصحية















 قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة
قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة "المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة
"المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة (نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)
(نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)


















