

النبات

مواضيع عامة في علم النبات

الجذور - السيقان - الأوراق

النباتات الوعائية واللاوعائية

البذور (مغطاة البذور - عاريات البذور)

الطحالب

النباتات الطبية


الحيوان

مواضيع عامة في علم الحيوان

علم التشريح

التنوع الإحيائي

البايلوجيا الخلوية


الأحياء المجهرية

البكتيريا

الفطريات

الطفيليات

الفايروسات


علم الأمراض

الاورام

الامراض الوراثية

الامراض المناعية

الامراض المدارية

اضطرابات الدورة الدموية

مواضيع عامة في علم الامراض

الحشرات


التقانة الإحيائية

مواضيع عامة في التقانة الإحيائية


التقنية الحيوية المكروبية

التقنية الحيوية والميكروبات

الفعاليات الحيوية

وراثة الاحياء المجهرية

تصنيف الاحياء المجهرية

الاحياء المجهرية في الطبيعة

أيض الاجهاد

التقنية الحيوية والبيئة

التقنية الحيوية والطب

التقنية الحيوية والزراعة

التقنية الحيوية والصناعة

التقنية الحيوية والطاقة

البحار والطحالب الصغيرة

عزل البروتين

هندسة الجينات


التقنية الحياتية النانوية

مفاهيم التقنية الحيوية النانوية

التراكيب النانوية والمجاهر المستخدمة في رؤيتها

تصنيع وتخليق المواد النانوية

تطبيقات التقنية النانوية والحيوية النانوية

الرقائق والمتحسسات الحيوية

المصفوفات المجهرية وحاسوب الدنا

اللقاحات

البيئة والتلوث


علم الأجنة

اعضاء التكاثر وتشكل الاعراس

الاخصاب

التشطر

العصيبة وتشكل الجسيدات

تشكل اللواحق الجنينية

تكون المعيدة وظهور الطبقات الجنينية

مقدمة لعلم الاجنة


الأحياء الجزيئي

مواضيع عامة في الاحياء الجزيئي


علم وظائف الأعضاء


الغدد

مواضيع عامة في الغدد

الغدد الصم و هرموناتها

الجسم تحت السريري

الغدة النخامية

الغدة الكظرية

الغدة التناسلية

الغدة الدرقية والجار الدرقية

الغدة البنكرياسية

الغدة الصنوبرية

مواضيع عامة في علم وظائف الاعضاء

الخلية الحيوانية

الجهاز العصبي

أعضاء الحس

الجهاز العضلي

السوائل الجسمية

الجهاز الدوري والليمف

الجهاز التنفسي

الجهاز الهضمي

الجهاز البولي


المضادات الميكروبية

مواضيع عامة في المضادات الميكروبية

مضادات البكتيريا

مضادات الفطريات

مضادات الطفيليات

مضادات الفايروسات

علم الخلية

الوراثة

الأحياء العامة

المناعة

التحليلات المرضية

الكيمياء الحيوية

مواضيع متنوعة أخرى

الانزيمات
Yeast GAL Genes: A Model for Activation and Repression
المؤلف:
JOCELYN E. KREBS, ELLIOTT S. GOLDSTEIN and STEPHEN T. KILPATRICK
المصدر:
LEWIN’S GENES XII
الجزء والصفحة:
3220
Yeast GAL Genes: A Model for Activation and Repression
KEY CONCEPTS
- GAL1/10 genes are positively regulated by the activator Gal4.
- Gal4 is negatively regulated by Gal80.
- Gal80 is negatively regulated by Gal3, the ultimate positive regulator, which is activated by the inducer, galactose.
- GAL1/10 genes are negatively regulated by a noncoding RNA synthesized from a cryptic promoter that controlschromatin structure.
- Activated Gal4 recruits the machinery necessary to alter the chromatin and recruit RNA polymerase.
- Catabolite repression is mediated by a glucosedependent protein kinase, Snf1.
Yeast, like bacteria, need to be able to rapidly respond to their environment (see the chapter titled The Operon). In the yeast Saccharomyces cerevisiae, the GAL genes serve a similar function to the lac operon in E. coli. In an emergency, when there is little or no glucose as an energy source and only galactose (or in E. coli, lactose) is available, the cell will survive because it can catabolize the alternate sugar to generate ATP. The GAL system in S. cerevisiae has been a model system to investigate gene regulation in eukaryotes for many years. This section focuses on two of these genes, GAL1 and GAL10, which are shown in FIGURE 1. Like most eukaryotic genes, the GAL genes are monocistronic. These two genes are divergently transcribed and regulated from a central control region called the upstream activating sequence (UAS), which is similar to an enhancer. Like the lac operon in E. coli, the GAL genes are induced by their substrate, galactose. For the same reason as in E. coli, the GAL genes are also under another level of control (described shortly)—catabolite repression. They cannot be activated by the substrate galactose when there is a sufficient supply of glucose, the preferred energy source.
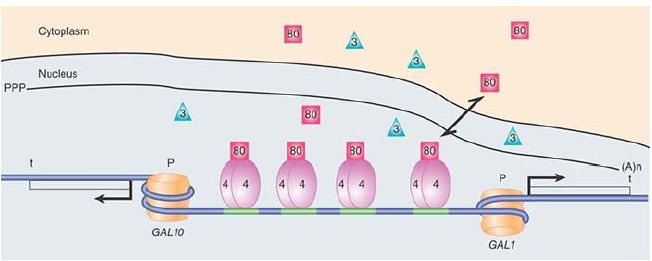
FIGURE 1. The yeast GAL1/GAL10 locus highlighting the UAS and showing the Gal4, Gal80, and Gal3 regulatory proteins and the RSC/nucleosome. Nucleosomes are also positioned at the promoters when the genes are not being transcribed.
Together, the GAL genes are under five different levels of control. The first level is chromatin structure. Mutations in any of the subunits of the chromatin remodeler SWI/SNF and in the acetyltransferase complex SAGA will result in reduced expression of the GAL genes. Second, the UAS has both general enhancer and Mig1 repressor–binding sites. The third level is through a noncoding RNA transcript that assists in maintaining repressed chromatin over the open reading frames. The fourth level is the GAL-specific galactose induction mechanism. The fifth level is catabolite (glucose) repression.
The two GAL genes are unusual in that they lack the typical nucleosome-free region present at the start sites of most yeast genes. Instead, the start sites are contained in well-positioned nucleosomes. The UAS region that controls the GAL genes has an unusual base composition—short-phased AT repeats every 10 base pairs—which causes the DNA to bend. Nucleosomes containing the histone variant H2AZ (Htz1 in yeast) are positioned over the promoters of both GAL1 and GAL10, aided in their positioning in part by the bent DNA.
The GAL10 gene is also an unusual gene in that it has a cryptic promoter in open chromatin at its 3′ end. This promoter transcribes a noncoding RNA that is antisense to GAL10 and extends through and includes GAL1 .
Transcription is very inefficient and the RNA abundance is extremely low (less than one copy per cell), due, in part, to rapid degradation. Under repressed conditions this promoter is stimulated by the Reb1 transcription factor, usually thought to be an RNA polymerase I transcription factor. The noncoding transcript represses transcription of the GAL1/10 pair of genes by recruiting the Set2 methyltransferase, which leads to H3K36 di- and trimethylation. H3K36me2/me3 recruits an HDAC to deacetylate the chromatin, which, in turn, leads to repressed chromatin structure.
The GAL genes are ultimately controlled by the positive regulator Gal4, which binds as a dimer to four binding sites in the UAS region, as shown in Figure 1and FIGURE 2. Its activation domain consists of two acidic patch domains. Gal4, in turn, is regulated by Gal80, a negative regulator that binds to Gal4 and masks its activation domain, preventing it from activating transcription. This is the normal state for the GAL genes: turned off and waiting to be induced. The chromatin architecture of the UAS has been difficult to discern. Recent data from uninduced cells suggest that a partly unwrapped nucleosome is constitutively held in place and positioned by the chromatin-remodeling factor RSC.
RSC in yeast, unlike its homologs in higher eukaryotes, has a domain for sequence-specific DNA binding. This complex facilitates the binding of Gal4 by aiding in the phasing of the nucleosomes over the two promoters and prevents them from encroaching on the Gal4 binding sites.
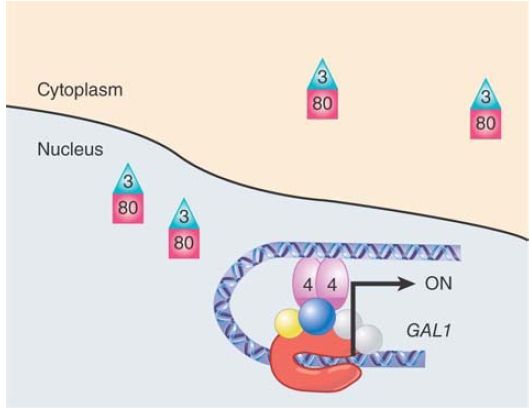
FIGURE 2. The yeast GAL1 gene as it is being activated. Gal3 is bound to Gal80 in the nucleus and cytoplasm, preventing it from binding to Gal4 and allowing Gal4 to recruit the transcription machinery and activate transcription.
Gal80, itself is regulated by the negative regulator Gal3, which is controlled by the inducer galactose. Gal80 contains overlapping binding sites for both Gal4 and Gal3. Gal3 is an interesting protein, having very high homology to Gal1, which is a galactokinase enzyme whose function is to phosphorylate galactose. Gal3 has no enzymatic activity, but retains the ability to bind galactose and ATP.
This changes the structure of Gal3 to enable it to bind to Gal80 in the presence of NADP. When it does, Gal3 masks the Gal4 binding site of Gal80, preventing it from binding to Gal4. This transition occurs very rapidly, leading to induction of Gal1/10, due primarily to Gal3 binding Gal80 in the nucleus. Gal3 is thus a negative regulator of a negative regulator, which makes it a positive regulator of Gal4.
This depletes the nuclear level of Gal80, unmasking Gal4 and allowing activation of the genes. NADP is thought to be a “second messenger” metabolic sensor.
Unmasked Gal4 is now able to begin the process of turning on the GAL1/10 genes through direct contact with a number of proteins at the promoter. During induction, Reb1 no longer binds to the cryptic promoter in GAL10. Gal4 recruits an H2B histone ubiquitylation factor (Rad6), which then stimulates histone di- and trimethylation of histone H3K4 by Set1. Next, the SAGA acetyltransferase complex is recruited by Gal4 and both deubiquitylates H2B and acetylates histone H3, ultimately resulting in the eviction of the poised nucleosomes from the two promoters. The removal is facilitated by the remodeler SWI/SNF and the chaperones Hsp90/70. SWI/SNF is not absolutely required but speeds up the process. This allows the recruitment of TBP/TF D, which then recruits RNA polymerase II and the coactivator complex Mediator.
Activated Gal4 directly contacts Mediator to ultimately initiate transcription. The elongation control factor TF S is also recruited, which actually plays a role in initiation for at least some genes. During the elongation phase of transcription, nucleosomes are disrupted . In order to prevent spurious transcription from internal cryptic promoters on either strand, histone octamers must re-form as RNA polymerase II passes. A number of histone chaperones and the FACT (facilitating chromatin transcription) complex play a role in the dynamics of octamer disassembly and assembly during elongation.
This system is also poised to rapidly repress transcription when the supply of galactose is used up or glucose becomes available. As Gal4 is activating transcription by RNA polymerase II, protein kinases associated with the activation of the polymerase also phosphorylate Gal4. This phosphorylation then leads to ubiquitination and destruction of Gal4. This turnover may be essential for RNA polymerase clearance and elongation. This is a dynamic system in which there must be a continuous positive signal, the presence of galactose.
Although catabolite repression in eukaryotes is used for the same purpose as in E. coli (which uses cAMP as a positive coregulator), it has a completely different mechanism. Glucose is a preferred sugar source compared to galactose. If the cell has both sugars, it will preferentially use the best source, glucose, and repress the genes for galactose utilization. Glucose repression of the yeast GAL genes is multifaceted. The glucose-dependent switch is the protein kinase Snf1. In low glucose, the GAL genes are transcribed because the general glucose-dependent repressor Mig1 has been inactivated, phosphorylated by Snf1. Glucose repression inactivates Snf1, which allows Mig1 to be active.
A number of other genes involving galactose usage are also downregulated in glucose, including the galactose transporter and Gal4 itself. Glucose inactivates Snf1, which leads to the activation of Mig1 at the GAL locus. Mig1 interacts at the GAL locus with the Cyc8-Tup1 corepressor, which is known to recruit histone deacetylases.
 الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
 اخر الاخبار
اخر الاخبار
اخبار العتبة العباسية المقدسة

الآخبار الصحية















 قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة
قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة "المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة
"المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة (نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)
(نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)


















