

النبات

مواضيع عامة في علم النبات

الجذور - السيقان - الأوراق

النباتات الوعائية واللاوعائية

البذور (مغطاة البذور - عاريات البذور)

الطحالب

النباتات الطبية


الحيوان

مواضيع عامة في علم الحيوان

علم التشريح

التنوع الإحيائي

البايلوجيا الخلوية


الأحياء المجهرية

البكتيريا

الفطريات

الطفيليات

الفايروسات


علم الأمراض

الاورام

الامراض الوراثية

الامراض المناعية

الامراض المدارية

اضطرابات الدورة الدموية

مواضيع عامة في علم الامراض

الحشرات


التقانة الإحيائية

مواضيع عامة في التقانة الإحيائية


التقنية الحيوية المكروبية

التقنية الحيوية والميكروبات

الفعاليات الحيوية

وراثة الاحياء المجهرية

تصنيف الاحياء المجهرية

الاحياء المجهرية في الطبيعة

أيض الاجهاد

التقنية الحيوية والبيئة

التقنية الحيوية والطب

التقنية الحيوية والزراعة

التقنية الحيوية والصناعة

التقنية الحيوية والطاقة

البحار والطحالب الصغيرة

عزل البروتين

هندسة الجينات


التقنية الحياتية النانوية

مفاهيم التقنية الحيوية النانوية

التراكيب النانوية والمجاهر المستخدمة في رؤيتها

تصنيع وتخليق المواد النانوية

تطبيقات التقنية النانوية والحيوية النانوية

الرقائق والمتحسسات الحيوية

المصفوفات المجهرية وحاسوب الدنا

اللقاحات

البيئة والتلوث


علم الأجنة

اعضاء التكاثر وتشكل الاعراس

الاخصاب

التشطر

العصيبة وتشكل الجسيدات

تشكل اللواحق الجنينية

تكون المعيدة وظهور الطبقات الجنينية

مقدمة لعلم الاجنة


الأحياء الجزيئي

مواضيع عامة في الاحياء الجزيئي


علم وظائف الأعضاء


الغدد

مواضيع عامة في الغدد

الغدد الصم و هرموناتها

الجسم تحت السريري

الغدة النخامية

الغدة الكظرية

الغدة التناسلية

الغدة الدرقية والجار الدرقية

الغدة البنكرياسية

الغدة الصنوبرية

مواضيع عامة في علم وظائف الاعضاء

الخلية الحيوانية

الجهاز العصبي

أعضاء الحس

الجهاز العضلي

السوائل الجسمية

الجهاز الدوري والليمف

الجهاز التنفسي

الجهاز الهضمي

الجهاز البولي


المضادات الميكروبية

مواضيع عامة في المضادات الميكروبية

مضادات البكتيريا

مضادات الفطريات

مضادات الطفيليات

مضادات الفايروسات

علم الخلية

الوراثة

الأحياء العامة

المناعة

التحليلات المرضية

الكيمياء الحيوية

مواضيع متنوعة أخرى

الانزيمات
Introduction to Eukaryotic Transcription Regulation
المؤلف:
JOCELYN E. KREBS, ELLIOTT S. GOLDSTEIN and STEPHEN T. KILPATRICK
المصدر:
LEWIN’S GENES XII
الجزء والصفحة:
9-6-2021
2370
Introduction to Eukaryotic Transcription Regulation
Key concept
- Eukaryotic gene expression is usually controlled at the level of initiation of transcription by opening the chromatin.
The phenotypic differences that distinguish the various kinds of cells in a higher eukaryote are largely due to differences in the expression of genes that code for proteins; that is, those transcribed by RNA polymerase II. In principle, the expression of these genes can be regulated at any one of several stages. FIGURE 26.1 distinguishes (at least) six potential control points,
which form the following series:
Activation of gene structure: open chromatin
↓
Initiation of transcription and elongation
↓
Processing the transcript
↓
Transport to the cytoplasm from the nucleus
↓
Translation of mRNA
↓
Degradation and turnover of mRNA
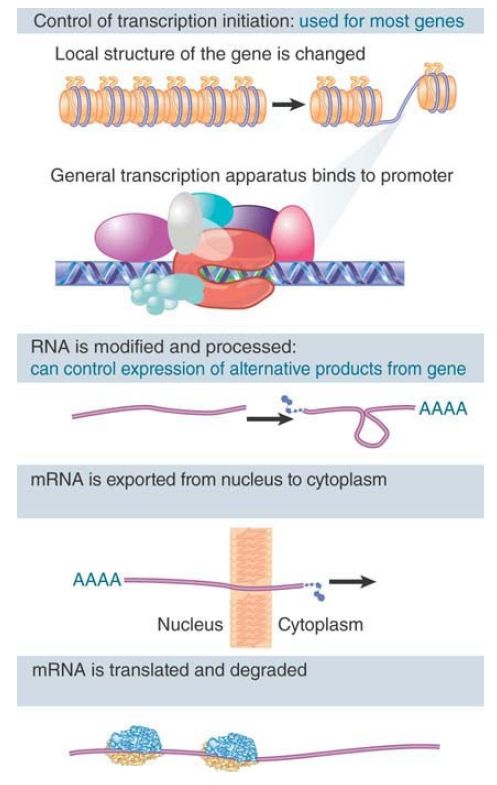
FIGURE 1. Gene expression is controlled principally at the initiation of transcription. Control of processing may be used to determine which form of a gene is represented in mRNA. The mRNA may be regulated during transport to the cytoplasm, during translation, and by degradation.
Whether a gene is expressed depends on the structure of chromatin both locally (at the promoter) and in the surrounding domain. Chromatin structure correspondingly can be regulated by individual activation events or by changes that affect a wide chromosomal region. The most localized events concern an individual target gene, where changes in nucleosomal structure and organization occur in the immediate vicinity of the promoter. Many genes have multiple promoters; the choice of the promoter can alter the pattern of regulation and influence how the mRNA is used because it will change the 5′ untranslated region (UTR). More general changes may affect regions as large as a whole chromosome. Activation of a gene requires changes in the state of chromatin. The essential issue is how the transcription factors gain access to the promoter DNA.
Local chromatin structure is an integral part of controlling gene expression. Broadly speaking, genes may exist in either of two basic structural conditions. The first is an inactive gene in closed chromatin. Alternatively, genes are found in an “active” state, or open chromatin, only in the cells in which they are expressed, or potentially expressed. The change of structure precedes the act of transcription and indicates that the gene is able to be transcribed.
This suggests that acquisition of the active structure must be the first step in gene expression. Active genes are typically found in domains of euchromatin with a preferential susceptibility to
nucleases, and hypersensitive sites are created at promoters before a gene is activated (see the Chromatin chapter). A gene that is in open chromatin may actually be active and be transcribed, or it may be potentially active and waiting for a subsequent signal, a condition called poised.
An intimate and continuing connection exists between initiation of transcription and chromatin structure. Some activators of gene transcription directly modify histones; in particular, acetylation of histones is associated with gene activation. Conversely, some repressors of transcription function by deacetylating histones.
Thus, a reversible change in histone structure in the vicinity of the promoter is involved in the control of gene expression. These changes influence the association of histone octamers with DNA and are responsible for controlling the presence and structure of nucleosomes at specific sites. This is an important aspect of the mechanism by which a gene is maintained in an active or inactive state.
The mechanisms by which regions of chromatin are maintained in an inactive (silent) state are related to the means by which an individual promoter is repressed. The proteins involved in the
formation of heterochromatin act on chromatin via the histones, and modifications of the histones are an important feature in the interaction. Once established, such changes in chromatin can persist through cell divisions, creating an epigenetic state in which the properties of a gene are determined by the self-perpetuating structure of chromatin. The name epigenetic reflects the fact that a gene may have an inherited condition (it may be active or inactive) that does not depend solely on its sequence . Once transcription begins, regulation during the elongation phase of transcription is also possible . However, attenuation, such as that in bacteria , cannot occur in eukaryotes because of the separation of chromosomes from the cytoplasm by the nuclear membrane. The primary mRNA transcript is modified by capping at the 5′ end and for most protein-coding genes is also modified by polyadenylation at the 3′ end (see the chapter RNA Splicing and Processing).
Many genes also have multiple termination sites, which can alter the 3′ UTR, and thus mRNA function and behavior. Introns must be excised from the transcripts of interrupted genes. The mature RNA must then be exported from the nucleus to the cytoplasm. Regulation of gene expression at the level of nuclear RNA processing might involve any or all of these stages, but the one that has the most evidence concerns changes in splicing; some genes are expressed by means of alternative splicing patterns whose regulation controls the type of protein product .
The translation of an mRNA in the cytoplasm can be specifically controlled, as can the turnover rate of the mRNA. This can also involve the localization of the mRNA to specific sites where it is expressed; in addition, the blocking of initiation of translation by specific protein factors may occur. Different mRNAs may have different intrinsic half-lives determined by specific sequence elements .
Regulation of tissue-specific gene transcription lies at the heart of eukaryotic differentiation. It is also important for control of metabolic and catabolic pathways. Gene regulators are typically
proteins; however, RNAs can also serve as gene regulators. This raises two questions about gene regulation:
- How does a protein transcription factor identify its group of target genes?
- How is the activity of the regulator itself regulated in response to intrinsic or extrinsic signals?
 الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
 اخر الاخبار
اخر الاخبار
اخبار العتبة العباسية المقدسة

الآخبار الصحية















 قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة
قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة "المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة
"المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة (نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)
(نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)


















