

النبات

مواضيع عامة في علم النبات

الجذور - السيقان - الأوراق

النباتات الوعائية واللاوعائية

البذور (مغطاة البذور - عاريات البذور)

الطحالب

النباتات الطبية


الحيوان

مواضيع عامة في علم الحيوان

علم التشريح

التنوع الإحيائي

البايلوجيا الخلوية


الأحياء المجهرية

البكتيريا

الفطريات

الطفيليات

الفايروسات


علم الأمراض

الاورام

الامراض الوراثية

الامراض المناعية

الامراض المدارية

اضطرابات الدورة الدموية

مواضيع عامة في علم الامراض

الحشرات


التقانة الإحيائية

مواضيع عامة في التقانة الإحيائية


التقنية الحيوية المكروبية

التقنية الحيوية والميكروبات

الفعاليات الحيوية

وراثة الاحياء المجهرية

تصنيف الاحياء المجهرية

الاحياء المجهرية في الطبيعة

أيض الاجهاد

التقنية الحيوية والبيئة

التقنية الحيوية والطب

التقنية الحيوية والزراعة

التقنية الحيوية والصناعة

التقنية الحيوية والطاقة

البحار والطحالب الصغيرة

عزل البروتين

هندسة الجينات


التقنية الحياتية النانوية

مفاهيم التقنية الحيوية النانوية

التراكيب النانوية والمجاهر المستخدمة في رؤيتها

تصنيع وتخليق المواد النانوية

تطبيقات التقنية النانوية والحيوية النانوية

الرقائق والمتحسسات الحيوية

المصفوفات المجهرية وحاسوب الدنا

اللقاحات

البيئة والتلوث


علم الأجنة

اعضاء التكاثر وتشكل الاعراس

الاخصاب

التشطر

العصيبة وتشكل الجسيدات

تشكل اللواحق الجنينية

تكون المعيدة وظهور الطبقات الجنينية

مقدمة لعلم الاجنة


الأحياء الجزيئي

مواضيع عامة في الاحياء الجزيئي


علم وظائف الأعضاء


الغدد

مواضيع عامة في الغدد

الغدد الصم و هرموناتها

الجسم تحت السريري

الغدة النخامية

الغدة الكظرية

الغدة التناسلية

الغدة الدرقية والجار الدرقية

الغدة البنكرياسية

الغدة الصنوبرية

مواضيع عامة في علم وظائف الاعضاء

الخلية الحيوانية

الجهاز العصبي

أعضاء الحس

الجهاز العضلي

السوائل الجسمية

الجهاز الدوري والليمف

الجهاز التنفسي

الجهاز الهضمي

الجهاز البولي


المضادات الميكروبية

مواضيع عامة في المضادات الميكروبية

مضادات البكتيريا

مضادات الفطريات

مضادات الطفيليات

مضادات الفايروسات

علم الخلية

الوراثة

الأحياء العامة

المناعة

التحليلات المرضية

الكيمياء الحيوية

مواضيع متنوعة أخرى

الانزيمات
DNA Repair in Eukaryotes Occurs in the Context of Chromatin
المؤلف:
JOCELYN E. KREBS, ELLIOTT S. GOLDSTEIN and STEPHEN T. KILPATRICK
المصدر:
LEWIN’S GENES XII
الجزء والصفحة:
21-4-2021
2752
DNA Repair in Eukaryotes Occurs in the Context of Chromatin
KEY CONCEPTS
- Both histone modification and chromatin remodeling are essential for repair of DNA damage in chromatin.
- H2A phosphorylation (γγ−H2AX) is a conserved DSBdependent modification that recruits chromatin-modifying activities and facilitates assembly of repair factors.
- Different patterns of histone modifications may distinguish stages of repair or different pathways of repair.
- Remodelers and chaperones are required to reset chromatin structure after completion of repair.
DNA repair in eukaryotic cells involves an additional layer of complexity: the nucleosomal packaging of the DNA substrate.
Chromatin presents an obstacle to DNA repair, as it does to replication and transcription, because nucleosomes must be displaced in order for processes such as strand unwinding, excision, or resection to occur. Chromatin in the vicinity of DNA damage must therefore be modified and remodeled before or during repair, and then the original chromatin state must be restored after repair is completed, as shown in FIGURE 1.

FIGURE 1. DNA damage in chromatin requires chromatin remodeling and histone modification for efficient repair; after repair the original chromatin structure must be restored.
Access to DNA in chromatin is controlled by a combination of covalent histone modifications, which change the structure of chromatin and create alternative binding sites for chromatin-binding proteins , and ATP-dependent chromatin remodeling , in which remodeling complexes use the energy of ATP to slide or displace nucleosomes. Both histone modification and chromatin remodeling have been implicated in all of the eukaryotic repair pathways discussed in this chapter; for example, both the global-genome and transcription-coupled pathways of nucleotide excision repair depend on specific chromatin-remodeling enzymes, and repair of UV-damaged DNA is facilitated by histone acetylation. A summary of the histone modifications implicated in different repair processes is shown in FIGURE 2. All four histones are modified in the course of double-strand break repair (discussed further below), and histone acetylation, methylation, phosphorylation, and ubiquitination at different sites are differentially involved in different repair pathways.
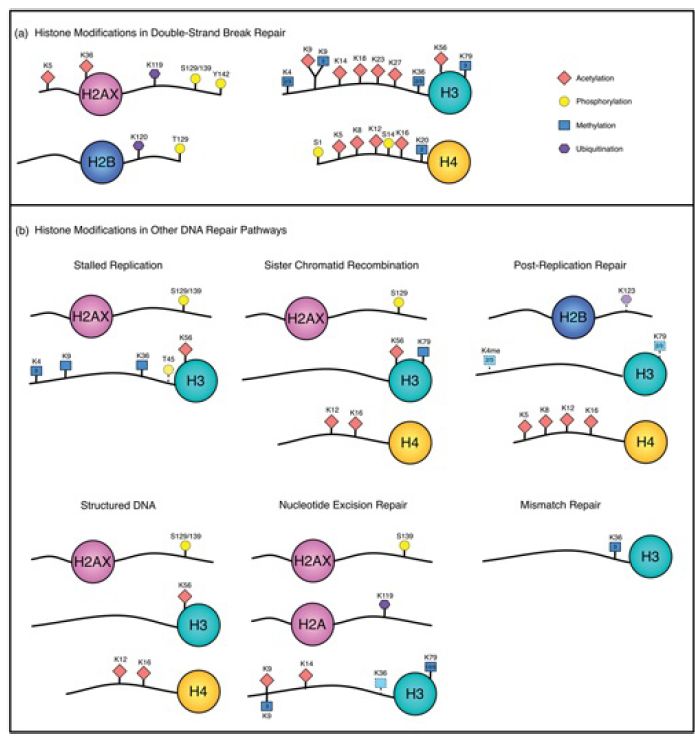
FIGURE 2. Histone modifications associated with different repair pathways. Histone phosphorylation (yellow circle), acetylation (red diamond), methylation (blue square), and ubiquitination (purple hexagon) have all been implicated in repair. Double-strand break repair (DSBR) is grouped as a single pathway, but certain modifications can be specific to different DSBR processes.
Figure generously provided by Nealia C. M. House and Catherine H. Freudenreich.
One of the most extensive posttranslational modifications that occurs following DNA damage (DSBs as well as other damage) in all eukaryotes examined except yeast is the poly-(ADP)- ribosylation (PARylation) of many histone and nonhistone targets.
This is catalyzed by enzymes in the poly-(ADP-ribose) polymerase (PARP) superfamily of NAD+ -dependent ADP-ribosyltransferases. PAR is a large, branched ADP-ribose polymer that is highly negatively charged, and in some cases the mass of PAR added to a protein can exceed the original mass of the unmodified target! One member of this family, PARP-1, auto-PARylates itself in response to DNA damage, which leads to its association with repair factors and their recruitment to sites of damage. The PARylation is turned over rapidly, and it is thought that this turnover is also important in the DNA damage response.
The best understanding of the roles of chromatin modification, however, is in the repair of DNA DSBs. Much of our understanding of the role of chromatin modification in double-strand break repair (DSBR) comes from studies in yeast utilizing a system derived from the yeast mating-type switching apparatus, which was introduced in the Homologous and Site-Specific Recombination chapter. In this experimental system, yeast strains contain a galactose-inducible
HO endonuclease, which generates a unique DSB at the active mating-type locus (MAT) when cells are grown in galactose. These breaks are repaired using the recombination-repair factors described in the section in this chapter titled Recombination-Repair of Double-Strand Breaks in Eukaryotes, using homologous sequences present at the silent mating-type loci HML or HMR. In the absence of homologous donor sequences (or, for haploid yeast, a sister chromatid during S/G2), cells utilize the second major pathway of DSB repair, NHEJ, to directly ligate broken chromosome ends.
Using this system (and other methods for inducing DSBs in mammalian systems as well), researchers have identified numerous histone modifications and chromatin-remodeling events that take place during repair. The best characterized of these is the phosphorylation of the histone H2AX variant . The major H2A in yeast is actually of the H2AX type, which is distinguished by an SQEL/Y motif at the end of the Cterminal tail. (This variant makes up only 5% to 15% of the total H2A in mammalian cells.) The serine in the SQEL/Y sequence is the substrate for phosphorylation by the Mec1/Tel1 kinases in yeast, homologs of the mammalian ATM/ATR kinases (ATM is the checkpoint kinase affected in AT patients, discussed in the previous section). H2AX phosphorylated at this site (serine 129 in yeast, 139 in mammals) is referred to as γ-H2AX.
γ-H2AX is a universal marker for DSBs in eukaryotes, whether they occur as a result of damage, or during their normal appearance during mating-type switching in yeast, or during meiotic recombination in numerous species. γ-H2AX phosphorylation is one of the earliest events to occur at a DSB, appearing close to the breakpoint within minutes of damage and spreading to include as much as 50 kb of chromatin in yeast and megabases of chromatin in mammals. γ-H2AX is detectable throughout the repair process and is linked to checkpoint recovery after repair. H2AX phosphorylation stabilizes the association of repair factors at the breakpoint and also serves to recruit chromatin-remodeling enzymes and a histone acetyltransferase to facilitate subsequent stages of repair.
In addition to γ-H2AX, numerous other histone modification events occur at DSBs at defined points during the repair process. Some of these are summarized in FIGURE 3., which shows an approximate timeline of modification events at an HO-induced break in yeast. They include transient phosphorylation of H4S1 by casein kinase 2, a modification more important for NHEJ than DSBR, and complex, asynchronous waves of acetylation of both histones H3 and H4 that are controlled by at least three different acetyltransferases and three different deacetylases. It has recently been shown that γ-H2AX is further subject to polyubiquitylation following its phosphorylation, and dephosphorylation of a tyrosine in γ-H2AX (Y142 in mammals) is also critical in the damage response. Certain other preexisting modifications, such as methylated H4K20 and H3K79, also appear to play a role, perhaps by being exposed only upon chromatin conformational changes that occur in response to other modification at a damage site. It is not fully understood how each modification promotes different steps in the repair process (and the details may differ between species), but it is important to note that the patterns of modification differ between homologous recombination and end-joining pathways,suggesting that these modifications may recruit factors specific for the different repair mechanisms.
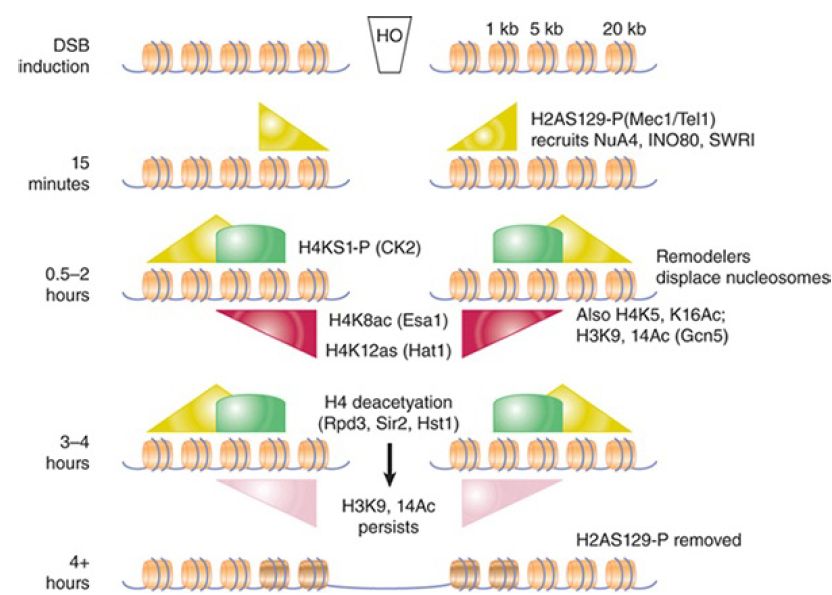
FIGURE 3.Summary of known histone modifications at an HOinduced double-strand break. The approximate timing of events is indicated on the left. Repair rates for homologous recombination and nonhomologous end joining differ in this experimental system, so the precise timing of different modification events relative to one another is not always directly comparable between pathways. The relative distances from the breakpoint are indicated in the upper right (not to scale). Shaded triangles and arcs show distributions and relative levels of the indicated modifications.
A number of chromatin-remodeling enzymes also act at DSBs. All chromatin-remodeling enzymes are members of the SWI2/SNF2 superfamily of enzymes, but there are numerous subfamilies within this group . At least three different subfamilies are implicated in DSBR: the SWI/SNF and RSC complexes of the SNF2 subfamily, the INO80 and SWR1 complexes of the INO80 group, and Rad54 and Rdh54 of the Rad54 subfamily. As discussed in the section in this chapter titled Recombination-Repair of Double-Strand Breaks in Eukaryotes, the Rad54 and Rdh54 enzymes play roles during the search for homologous donors and strand-invasion stages of repair, but other chromatin remodelers appear important during every stage, including initial damage recognition, strand resection, and the resetting of chromatin as repair is completed. This final stage also requires the activities of the histone chaperones Asf1 and CAF-1, which are needed to restore chromatin structure on the newly repaired region and allow recovery from the DNA damage checkpoint.
 الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
الاكثر قراءة في مواضيع عامة في الاحياء الجزيئي
 اخر الاخبار
اخر الاخبار
اخبار العتبة العباسية المقدسة

الآخبار الصحية















 قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة
قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة "المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة
"المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة (نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)
(نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)


















