

النبات

مواضيع عامة في علم النبات

الجذور - السيقان - الأوراق

النباتات الوعائية واللاوعائية

البذور (مغطاة البذور - عاريات البذور)

الطحالب

النباتات الطبية


الحيوان

مواضيع عامة في علم الحيوان

علم التشريح

التنوع الإحيائي

البايلوجيا الخلوية


الأحياء المجهرية

البكتيريا

الفطريات

الطفيليات

الفايروسات


علم الأمراض

الاورام

الامراض الوراثية

الامراض المناعية

الامراض المدارية

اضطرابات الدورة الدموية

مواضيع عامة في علم الامراض

الحشرات


التقانة الإحيائية

مواضيع عامة في التقانة الإحيائية


التقنية الحيوية المكروبية

التقنية الحيوية والميكروبات

الفعاليات الحيوية

وراثة الاحياء المجهرية

تصنيف الاحياء المجهرية

الاحياء المجهرية في الطبيعة

أيض الاجهاد

التقنية الحيوية والبيئة

التقنية الحيوية والطب

التقنية الحيوية والزراعة

التقنية الحيوية والصناعة

التقنية الحيوية والطاقة

البحار والطحالب الصغيرة

عزل البروتين

هندسة الجينات


التقنية الحياتية النانوية

مفاهيم التقنية الحيوية النانوية

التراكيب النانوية والمجاهر المستخدمة في رؤيتها

تصنيع وتخليق المواد النانوية

تطبيقات التقنية النانوية والحيوية النانوية

الرقائق والمتحسسات الحيوية

المصفوفات المجهرية وحاسوب الدنا

اللقاحات

البيئة والتلوث


علم الأجنة

اعضاء التكاثر وتشكل الاعراس

الاخصاب

التشطر

العصيبة وتشكل الجسيدات

تشكل اللواحق الجنينية

تكون المعيدة وظهور الطبقات الجنينية

مقدمة لعلم الاجنة


الأحياء الجزيئي

مواضيع عامة في الاحياء الجزيئي


علم وظائف الأعضاء


الغدد

مواضيع عامة في الغدد

الغدد الصم و هرموناتها

الجسم تحت السريري

الغدة النخامية

الغدة الكظرية

الغدة التناسلية

الغدة الدرقية والجار الدرقية

الغدة البنكرياسية

الغدة الصنوبرية

مواضيع عامة في علم وظائف الاعضاء

الخلية الحيوانية

الجهاز العصبي

أعضاء الحس

الجهاز العضلي

السوائل الجسمية

الجهاز الدوري والليمف

الجهاز التنفسي

الجهاز الهضمي

الجهاز البولي


المضادات الميكروبية

مواضيع عامة في المضادات الميكروبية

مضادات البكتيريا

مضادات الفطريات

مضادات الطفيليات

مضادات الفايروسات

علم الخلية

الوراثة

الأحياء العامة

المناعة

التحليلات المرضية

الكيمياء الحيوية

مواضيع متنوعة أخرى

الانزيمات
Ethanol Production from Biomass
المؤلف:
John M Walker and Ralph Rapley
المصدر:
Molecular Biology and Biotechnology 5th Edition
الجزء والصفحة:
26-1-2021
3154
Ethanol Production from Biomass
SHF and SSF Processing
Enzyme manufacture for SHF and SSF/SSCF applications have grown in anticipation of biomass ethanol biorefineries that will require cellulases and other biomass enzymes. An important producer of cellulases for biomass hydrolysis is Trichoderma reesei and Genencor/Danisco has used microarray technology to evaluate and presumably improve the enzyme production. Similar approaches are undoubtedly underway at Novozymes and other enzyme manufacturers, but much will remain unavailable until patent application publication. Target enzymes include 1,4-β-cellobiosidases and endo-1,4-β-D-xylanases. Indeed, Wilson has
used microarray and proteomic analysis to detect induction of a xylanase with cellobiose by a biomass degrading bacterium T. fusca.
Finally, in the Fall of 2007, Genencor launched a new enzyme formulation intended for SSF conversion, called Accellerase, which is a blend of cellulases and cellobiosidases (genencor.com). The process for production of ethanol from biomass described earlier contains two biological unit operations: enzymatic hydrolysis of the complex biomass carbohydrates cellulose and hemicellulose that remain after pretreatment by exogenously added enzymes, and fermentation of the resulting simple carbohydrates by an ethanologenic microorganism to ethanol. The latter fermentation operation can comprise two fermentations: conversion of cellulose-derived glucose to ethanol and fermentation of hemicellulose-derived pentose/xylose sugars to ethanol.
Depending on the fermentation capabilities of the ethanologenic microorganism chosen, either one or two fermentation steps are required,since many natural ethanol-producing microorganisms such as S. cerevisiae do not ferment xylose/pentose sugars.
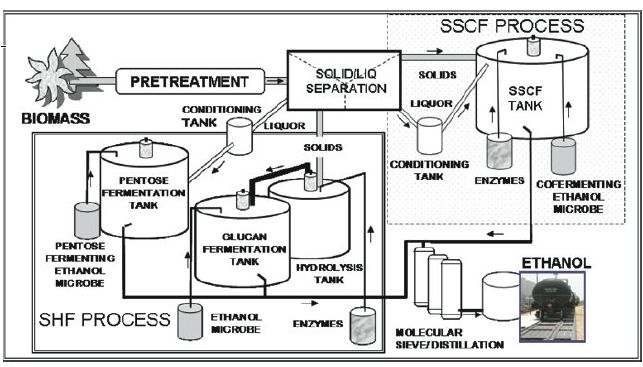
Figure .1 Diagram of biomass fermentation by either the separate hydrolysis and fermentation process (SHF) (left) or simultaneous saccharification and cofermentation (SSCF) process (right). SHF also shows conversion of glucan solids and xylose hydrolyzates separately with different ethanolproducing microorganisms, whereas SSCF requires an organism able to ferment both carbohydrate sources. Use of a co-fermenting microorganism in the SHF process will reduce the tank number by one. Conditioning is a process to remove fermentation inhibitors from the pretreatment liquor. Waste streams not included.
As described earlier, separate hydrolysis and fermentation operations (SHF) permit the rapid
enzymatic hydrolysis of cellulose at elevated temperatures (B60 1C), followed by free sugar fermentation to ethanol at a lower temperature.
As shown in Figure 1, this process has been largely replaced in typical plant designs by the simpler SSF process, in which cellulose hydrolysis and ethanol production are conducted in one vessel. The ability to use only one vessel requires the use of an ethanologic microorganism that can ferment hexose and pentose sugars to avoid the double vessel configuration shown in the SHF schematic in Figure .1. Note the use of a conditioning vessel that is needed to remove fermentation inhibitors contributed by the pretreatment process, which will be discussed further below. This technology, in various configurations, is the state-of-art heading to commercialization. Indeed, a pilot plant has been in operation at the 660 000 gallons per year scale at Iogen Corporation in Ottawa, Canada, using SSF technology. Still no full-scale plant is in operation at present but a number are planned, including some of the action of the cellulases (endogluconase, exogluconases) proceeds, significant levels of the cellobiose are produced which are known to competitively inhibit the cellulase action. As a result of this, commercial cellulase preparations, such as Accellerase, used in the SSF process include extra b-glucosidase. Improper enzyme ratios, differential inactivation or overall insufficient enzyme addition could result in diminished sugar availability during the SSF process, producing a carbon- limited fermentation. Like the SHF process, both transcriptomic and metabolomic analysis should be able to detect depleted carbon flow by initial starvation response accompanied by decreased sugar intermediates in the metabolome. The gene response during carbon source depletion will likely be accompanied by increases in expression of signal transduction and sugar transport genes, for example, as genetic indicationsof hydrolytic imbalance, as has been seen with pure sugar fermentations.
If the ethanologen is capable of fermenting xylose and arabinose in SSCF mode, an additional level of complexity exists that can readily be evaluated by transcriptomic analysis of the pentose phosphate pathway during the SSF conversion. This examination may be particularly valuable if the xylose utilization capability is the result of addition of genetic modification,including possible addition of exogenous foreign genes involved in pentose sugar conversion. In this regard, detection of the level of a exogenous enzyme, such as the anaerobic fungal xylose isomerase, could be readily detected by both transcriptomic and proteomic analysis permitting estimation of whether the fermentation conditions supports sufficient levels of this or other critical enzymes throughout the SSF process.
Complete utilization of the pentose sugars (primarily xylose and arabinose) has been a vexing problem not easily accomplished with both yeast and some bacterial that negatively impacts SSF fermentations. The pentose phosphate pathway (PPP) is very complex and this common but unexplainable incomplete fermentation may be the result of an imbalance of carbon flow, redox balance or net energy balance. Indeed, fermentation of high levels of glucose and xylose (50:50) by S. cerevisiae preferentially converted to xylose to xylitol and not ethanol after producing ethanol from glucose.Determination of the pentose phosphate pathway and key glycolytic intermediates showed that increased xylose conversion was the result of higher levels of PPP intermediates, but whether this was due to flux rates or downstream pathway limitations could not be determined. A similar approach is needed for the more complex SSF fermentation during initial rapid pentose sugar fermentation compared with later during the fermentation where the pentose conversion wanes and often is not complete. Such analysis is likely to detect PPP gene expression changes, depletion of PPP enzymes in the proteome and particularly changes in the contents of the metabolome or fluxome as the pentose sugar fermentation process changes during the SSF process. Detection of the cause of the metabolic imbalance preventing full conversion of the pentose sugars would permit a directed genetic and/or fermentation engineering solution to be selected and improvements made in S. cerevisiae, E. coli and Z.mobilis ethanologens developed over the last decade.
Impact of Enzyme and Fermentation Inhibitors
Fermentation for beer, wine and biomass ethanol does not benefit from a relatively pure source of simple carbohydrates that Brazilian sugar cane production enjoys. These feedstocks contain a variety of soluble plant (flavor) components created during the processing. Wine fermentation contains limited fermentation inhibitors especially with white wine fermentation and additional inhibitors are common in beer production due to the cooking steps and the addition of hops. Neither of these processes approach the level of fermentation inhibitors present in the very complex milieu of partially hydrolyzed plant material after biomass has been pretreated, neutralized and conditioned prior to either SHF or more commonly SSF conversion. Among the most common inhibitory compounds for biomass conversion is free acetic acid liberated from acetylated hemicellulose during pretreatment. This acetate can reach typically ~2–4 gL-1 levels depending upon the plant substrate involved, providing a significant source of fermentation inhibition.
An additional source of inhibitors is the degradative products from lignin, which is a complex polyphenolic structural compound found in all plant matter.Acidic processing of biomass can yield aromatic acids, aldehydes, ketones and other derivatives, all of which are highly inhibitory to metabolic activity.Indeed, these compounds impact both cell growth and final ethanol concentration. While phenolic compounds from lignin breakdown provide significant inhibition to ethanologen’s metabolic activity, lignin itself, in various forms after pretreatment, has been implicated in reducing the enzyme activities needed for either successful SHF or SSF production of ethanol. It has been hypothesized that lignin is partially solubilized by both alkaline and acidic pretreatment at elevated temperatures and then it is redeposited on the biomass upon cooling. This has been confirmed with the development of a protein/enzyme protection process using low-cost protein that minimizes lignin inactivation or binding to cellulase enzymes needed for cellulose hydrolysis. Therefore, lignin poses a significant problem to biomass ethanol production, whether degraded or intact.
Dilute acid pretreatment and steam or hot water pretreatment result in an acidic environment during the initial biomass processing steps, whether from added acids or self-generated acetic acid from hemicellulose deacetylation. It is known that acid attack on sugars leads to the production of degradation products called furans, with xylose yielding furfural and glucose yielding hydroxymethylfurfural. This chemical reaction is accelerated at the high temperatures needed for effective pretreatment.
These furan derivatives not only emanate from monomeric sugars, but also kinetic analysis has determined that intact cellulose and cellulose oligomers are subject to acid attack generating furan inhibitors.
Regardless of the source, the pretreatment process must be operated carefully to minimize the excessive acidic hydrolysis of both the hemicellulose and cellulose since these furans are highly toxic to all ethanologenic microorganisms tested.Fortunately, there are a number of processes, collectively called conditioning (Figure .1), that can be used to minimize or eliminate the toxic byproducts of pretreatment of biomass, which include the over-liming process, that permit fermentation processes to proceed with a variety of ethanologen microorganisms.
Addition of fermentation inhibitors during ethanol production is needed to elucidate the metabolic impact of the myriad of chemical inhibitors, each of which may have a different impact on the fermentation process. Transcriptomic analysis before and immediately after charging fermentation with the selected inhibitor would be a powerful approach to detecting directly the impact of that inhibitor on the biological processes that occur during the fermentation. Work is under way at the USDA laboratory in Peoria, IL, to investigate the impact of furfural and hydroxyfurfurals on S. cerevisiae fermentations and has resulting in the
detection of specific genes associated with the pentose phosphate pathway for xylose utilization that mitigates the inhibitory response. Their results using microarray analysis indicate that there is a nontransitory multigene response to furfural addition that eventually leads to adaptation to this potent metabolic inhibitor and provide ample opportunities for further evaluation of a very complex genetic response.
Consolidated Bioprocessing
A process for production of biomass ethanol without addition of exogenous cellulase enzymes has been under development for years using mesophilic and thermophilic anaerobes capable of growing and fermenting cellulose to ethanol and other fermentative byproducts. This process is called consolidated bioprocessing (CBP) due to the elimination of the requirement for exogenously added enzymes as needed by the SHS or SSF processes.35 CBP process relies on the production of carbohydrase enzymes by numerous anaerobic bacteria that can grow on complex carbohydrates such as cellulose and hemicellulose. In particular, certain clostridia produce a complex carbohydrase enzyme complex, the aforementioned cellulosomes that are expressed outside the cell but remain attached. These enzyme complexes attach to cellulose using a cellulose-binding module, which facilitates bringing the cell in close proximity to the substrate and permitting hydrolysis and consumption of the cellulose and other complex carbohydrates. These cellulosomes are extremely complex and have been the subject of decades of research, especially with C. thermocellum. The research has centered on structural biology studies for many of these years and recent genome sequencing has augmented research considerably with the discovery of over 70 genes potentially associated with the cellulosome.40 However, research has been slowed by the lack of widely available genetic transfer systems.
Still, systems biology tools such as proteomic analysis can be critical to evaluating changes in cellulosome structure and composition both with and without genetic modification. For example, research has evaluated the expression of cellulosomal protein during growth on cellobiose, which does not require cellulosomal activity.Stevenson and Weimer identified 17 genes differentially expressed during growth on cellobiose or cellulose in continuous culture, six involved in carbohydrate metabolism, and observed the majority had higher differential expression, but no direct correlation was possible based upon growth rate or fermentation products. Doi’s group has investigated the response of selected cellulosomal enzymes for Clostridium cellulovorans to growth on a variety of substrates such as hemicellulose, cellulose and pectin carbohydrates and analyzed the enzyme composition of the cellulosome by RNA Northern blots and PAGE and discovered changes in the enzyme profile of known carbohydrase enzymes. Recently, Gold and Martin used metabolic labeling with C. thermocellum to identify changes in cellulosome protein subunit during growth on either cellobiose or cellulose using proteomic analysis with mass spectrometry.
They identified 16 new catalytic units but more than 23 putative cellulosomal proteins remained undetectable when grown on cellulose or cellobiose alone.
Recent work in our laboratory has investigated the linkage of complex carbohydrate growth substrate and expression of cellulosomal catalytic subunits using mass spectrometry-based proteomic analysis (to be published). C. thermocellum ATCC 27405 was grown in small fermenters with a variety of carbon sources including crystalline and amorphous cellulose, cellobiose and with various fixed mixtures of these materials with hemicellulose and/or pectin. These experiments were conducted with N metabolic labeling and multi-dimensional LC–MS/MS proteomic analysis. Interesting patterns of expression of endogluconases were observed with fermentations of crystalline and amorphous cellulose and cellobiose at the individual enzyme family level. Xylanase enzyme expression was poorly responsive to cellulose and increased in expression in the presence of cellobiose. Not surprisingly, since C. thermocellum does not metabolize xylose and other pentose sugars, xylanase enzymes showed no specific pattern of induction in the absence of cellulose. However, more than 21 additional cellulosomal proteins were detected and specific responses to alternative non-cellulose substrates were observed. This study forms the foundation for the analysis of the cellulosomal
composition present during the fermentation of natural biomass substrates such as switchgrass and poplar that are the subject of on-going research.
Genetic transformation systems have been developed for specific microorganisms capable of CBP, which opens heretofore closed doors for strain development aimed at the cellulosome and other metabolic processes. For example, genetic systems have been developed for a limited number of anaerobic cellulose-hydrolyzing microorganisms including Clostridium cellulolyticum, C. thermocellum and Ther- moanaerobacterium saccharolyticum.These genetic tool developments position the researchers to investigate the structure–function relationship of cellulosome gene components and the intricacies of the cellulose fermentation. An example of the power of a genetic system is seen with the recent genetic engineering of T. saccharolyticum into a homoethanologenic bacterium.This thermophilic anaerobe ferments preferentially hemicellulose through the action of its portfolio of hemicellulases and produce by fermentation ethanol, lactic acid, acetic acid and hydrogen. Genetic engineering strategies were used specifically to block key genes in the acetate and lactate pathways, thus terminating the production of these common fermentative organic acids. Through successive genetic blocking strategies, lactic acid production was eliminate first, followed by disruption of the acetate pathway, with an industrially important compensating increasing in ethanol production.With the elimination of organic acid production, this genetically developed strain of T. saccharolyticum has the potential to be developed into a useful microorganism for conversion of the hemicellulose portion of biomass that is particularly present in the alkaline pretreatment methods due to the lack of acid-catalyzed hemicellulose depolymerization. Development of a holistic picture of a biological system requires that gene and protein expression be examined and analyzed. This can be further connected to any resulting metabolic products, either extracellular as is common for fermentation processes or for intracellular metabolites, both typically detected by advance mass spectrometric techniques. This combined approach has recently been applied to the complex process of fermentation of cellulose or cellobiose to metabolic products (ethanol, acetate and lactate) in our laboratory. The experimental approach was the fermentation of either cellulose or cellobiose by C. thermocellum using two identical fermenters for dual biological replicates. Fermentations were sampled throughout exponential and stationary phase to the point of full carbon source depletion.
The transcriptomic analysis was conducted using the complete genome sequence that was completed by the US DOE Joint Genome Institute (JGI) (http://genome.jgi-psf.org/finished_microbes/cloth/cloth. home.html) using microarrays prepared at ORNL with 70-mer oligonucleotides for predicted ORFs and employing differential dye tagging as standard protocols.114 Typical fermentations yielded six sample points, so the differential microarray analysis was conducted in a variety of ways both within a single type of fermentation (cellobiose or cellulose) during exponential versus stationary growth or between fermentations (cellobiose versus cellulose). Analysis of the differential expression between cellobiose and cellulose fermentations yielded numerous differences, which included a strong induction of cellulosomal proteins as the culture reached stationary phase regardless of carbon
source (to be published). Both cultures supported higher levels of energy and biosynthetic pathways as expected for a rapidly growing culture.
Analysis between different carbon source cultures at either late exponential or stationary phase showed strong induction of cellulosomal proteins during active growth that tapered upon reaching the stationary phase. Although these are global analyses based upon functional groups, more detailed comparisons are under way with this total transcriptomic analysis. One approach that assists in this further analysis is cluster analysis for gene expression during six time points within fermentation. Cluster analysis for the cellulose fermentation permitted both clustering based on similar temporal comparisons and multiple categories of gene patters were observed with about 378 genes showing near level expression, while 192 genes showed significant decreasing expression throughout the fermentation. By comparison, 424 genes showed increasing genes expression throughout the fermentation process and 145 genes showed dramatic increased expression only upon reaching late stationary. Interestingly, only 11 of the expressed genes within this latter set were from the multi-gene sporulation pathway which contains 51 identified genes involved in sporulation. This is apparently because the minimal medium used for our research does not support sporulation of C. thermocellum (to be published).
Global or even functional group analysis provides information on general trends but analysis of specific gene patterns found within the huge database that microarrays provide can investigate specific questions. Among the logical targets for C. thermocellum are genes that impact ethanol production. C. thermocellum produces ethanol and organic acids acetate and lactate during fermentation, but these latter metabolites detract from the ethanol yield. Elimination of these organic acids by classical genetic engineering would be the preferred approach.
Examination of the genome of C. thermocellum detected two structural genes for lactate dehydrogenase (LDH) in the pathway to lactic acid production. One of these genes has been cloned from C. thermocellum ATCC 27405. It is not known if they were functional duplications or spurious crossovers, but knowledge of the expression of both genes would likely simplify the genetic engineering approach. Examination of the genetic expression of LDH by microarray analysis showed that indeed there was a significant difference in expression of two genes. One LDH gene (Genome ID Cthe0345) showed continuous temporal
expression in cells grown on cellulose, whereas the other (Genome ID Cthe1053) did not demonstrate any significant expression under the conditions used. Apparently only one gene is strongly expressed and should be the initial target for genetic modification or knockout. Interestingly, the gene that was cloned by O¨ zkan et al. was Cthe1053 and it demonstrated LDH activity when put under expression of the lac promoter. Either the gene regulatory signals for the Cthe1053 were faulty or the regulatory signals prevented expression of this otherwise functional Cthe1053 LDH gene under the conditions used at a high enough level to be detected by the microarray.
In additional to gene expression patterns determined with microarrays, examination of the proteins present in cells for different conditions or different strains can shed additional light on the cell’s metabolism. An important characteristic needed for C. thermocellum is tolerance to ethanol. The wild-type C. thermocellum is tolerant to less than 1% ethanol, but early researchers found that the microorganism can adapt to increasing ethanol added to the selection culture and have demonstrated growth in 2% ethanol.Strobel and co-workers have developed a strain of C. thermocellum that can grow in as high at 7% ethanol and used proteomic analysis to detect protein differences between the wild-type and the stable adapted strains.The study
detected numerous differentially expressed membrane proteins that were down-regulated in the ethanol-adapted strain, providing potential targets for genetic improvement by protein overexpression to introduce tolerance in potential production strains. Due to the high homology of Nature, it highly likely that genes that cause ethanol tolerance will function in related microorganisms for an analogous homologous gene, for example in T. saccharolyticum. Also, selection of production strains resistant to inhibitors such as acetate, furfural and lignin monomers remains an important potential avenue for strain improvement.
Gasification Ethanol Production
One of the most versatile processes for the utilization of carbonaceous material is gasification, where the carbonaceous feedstock is converted to CO, CO2, H2 and H2O. A gasifier can convert plant matter (biomass), plastics and rubber to these compounds due to the high temperature controlled anaerobic process. Interestingly, there are a group of anaerobic bacteria, some thermophiles, which are able to convert these gaseous compounds to cellular material and ethanol as a byproduct of fermentation.The biochemistry uses the reductive acetyl-CoA pathway to permit conversion of the synthesis gas to ethanol, acetate and other chemical byproducts. The process uses high-density Clostridium ljungdahlii as the microbial catalyst and, depending on the fermentation conditions, either acetic acid or ethanol has been produced.Indeed, production of acetic acid as a fermentation product has limited the yield of ethanol under certain fermentation conditions. The process requires gas to liquid to microorganism transfer, so improving mass transfer has been the primary target of process improvement over the past 20 years. However, claims of up to 80% consumption of gases in a
specially designed fermenter have been made by BRI Engineering, allowing potential commercialization of this process.
The potential for process improvement using genetic engineering of the key enzymes acetyl-CoA synthase and CO dehydrogenase could yield significant process improvements. Additionally, transcriptomic and proteomic analysis of wild-type strains during syngas fermentation would likely detect the level of expression of enzymes involved in ethanol synthesis and acetate production, shedding light on the importance of blocking acetate production. These include alcohol dehydrogenase for ethanol synthesis and acetate kinase and phosphotransacetylase for acetate production. Additionally, quantitative proteomics could detect potential pathway limitations, providing guidance for genetic engineering over expression of both predictable and unexpected proteins needed for syngas fermentation. To accomplish this, the genome sequence of C. ljungdahlii is required. This microorganism is in the pipeline for genome sequencing by the DOE JGI and has been under way in Gottschalk’s laboratory funded by Celanese Chemicals, so it remains to be seen if this genome sequence will be available soon.
 الاكثر قراءة في مواضيع عامة في التقانة الإحيائية
الاكثر قراءة في مواضيع عامة في التقانة الإحيائية
 اخر الاخبار
اخر الاخبار
اخبار العتبة العباسية المقدسة

الآخبار الصحية















 قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة
قسم الشؤون الفكرية يصدر كتاباً يوثق تاريخ السدانة في العتبة العباسية المقدسة "المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة
"المهمة".. إصدار قصصي يوثّق القصص الفائزة في مسابقة فتوى الدفاع المقدسة للقصة القصيرة (نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)
(نوافذ).. إصدار أدبي يوثق القصص الفائزة في مسابقة الإمام العسكري (عليه السلام)


















